自定义回归模型¶
与单变量模型一样,可以创建自己的自定义参数生存模型。为什么你可能想要这样做?
使用已知的概率分布创建新的/扩展AFT模型
使用关于主题的领域知识创建一个分段模型
迭代并拟合一个更精确的参数模型
lifelines 有一个非常简单的 API 来创建自定义参数回归模型。你只需要定义累积风险函数。例如,恒定风险回归模型的累积风险如下所示:
\[\begin{split}H(t, x) = \frac{t}{\lambda(x)}\\ \lambda(x) = \exp{(\vec{\beta} \cdot \vec{x}^{\,T})}\end{split}\]
其中 \(\beta\) 是我们要优化的未知数。
以下是一些自定义模型的示例。
[1]:
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
from lifelines.fitters import ParametricRegressionFitter
from autograd import numpy as np
from lifelines.datasets import load_rossi
class ExponentialAFTFitter(ParametricRegressionFitter):
# this class property is necessary, and should always be a non-empty list of strings.
_fitted_parameter_names = ['lambda_']
def _cumulative_hazard(self, params, t, Xs):
# params is a dictionary that maps unknown parameters to a numpy vector.
# Xs is a dictionary that maps unknown parameters to a numpy 2d array
beta = params['lambda_']
X = Xs['lambda_']
lambda_ = np.exp(np.dot(X, beta))
return t / lambda_
rossi = load_rossi()
# the below variables maps {dataframe columns, formulas} to parameters
regressors = {
# could also be: 'lambda_': rossi.columns.difference(['week', 'arrest'])
'lambda_': "age + fin + mar + paro + prio + race + wexp + 1"
}
eaf = ExponentialAFTFitter().fit(rossi, 'week', 'arrest', regressors=regressors)
eaf.print_summary()
| model | lifelines.ExponentialAFTFitter |
|---|---|
| duration col | 'week' |
| event col | 'arrest' |
| number of observations | 432 |
| number of events observed | 114 |
| log-likelihood | -686.37 |
| time fit was run | 2020-07-26 22:06:42 UTC |
| coef | exp(coef) | se(coef) | coef lower 95% | coef upper 95% | exp(coef) lower 95% | exp(coef) upper 95% | z | p | -log2(p) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| param | covariate | ||||||||||
| lambda_ | Intercept | 4.05 | 57.44 | 0.59 | 2.90 | 5.20 | 18.21 | 181.15 | 6.91 | <0.005 | 37.61 |
| age | 0.06 | 1.06 | 0.02 | 0.01 | 0.10 | 1.01 | 1.10 | 2.55 | 0.01 | 6.52 | |
| fin | 0.37 | 1.44 | 0.19 | -0.01 | 0.74 | 0.99 | 2.10 | 1.92 | 0.06 | 4.18 | |
| mar | 0.43 | 1.53 | 0.38 | -0.32 | 1.17 | 0.73 | 3.24 | 1.12 | 0.26 | 1.93 | |
| paro | 0.08 | 1.09 | 0.20 | -0.30 | 0.47 | 0.74 | 1.59 | 0.42 | 0.67 | 0.57 | |
| prio | -0.09 | 0.92 | 0.03 | -0.14 | -0.03 | 0.87 | 0.97 | -3.03 | <0.005 | 8.65 | |
| race | -0.30 | 0.74 | 0.31 | -0.91 | 0.30 | 0.40 | 1.35 | -0.99 | 0.32 | 1.63 | |
| wexp | 0.15 | 1.16 | 0.21 | -0.27 | 0.56 | 0.76 | 1.75 | 0.69 | 0.49 | 1.03 |
| AIC | 1388.73 |
|---|---|
| log-likelihood ratio test | 31.22 on 7 df |
| -log2(p) of ll-ratio test | 14.11 |
[2]:
class DependentCompetingRisksHazard(ParametricRegressionFitter):
"""
Reference
--------------
Frees and Valdez, UNDERSTANDING RELATIONSHIPS USING COPULAS
"""
_fitted_parameter_names = ["lambda1", "rho1", "lambda2", "rho2", "alpha"]
def _cumulative_hazard(self, params, T, Xs):
lambda1 = np.exp(np.dot(Xs["lambda1"], params["lambda1"]))
lambda2 = np.exp(np.dot(Xs["lambda2"], params["lambda2"]))
rho2 = np.exp(np.dot(Xs["rho2"], params["rho2"]))
rho1 = np.exp(np.dot(Xs["rho1"], params["rho1"]))
alpha = np.exp(np.dot(Xs["alpha"], params["alpha"]))
return ((T / lambda1) ** rho1 + (T / lambda2) ** rho2) ** alpha
fitter = DependentCompetingRisksHazard(penalizer=0.1)
rossi = load_rossi()
rossi["week"] = rossi["week"] / rossi["week"].max() # scaling often helps with convergence
covariates = {
"lambda1": rossi.columns.difference(['week', 'arrest']),
"lambda2": rossi.columns.difference(['week', 'arrest']),
"rho1": "1",
"rho2": "1",
"alpha": "1",
}
fitter.fit(rossi, "week", event_col="arrest", regressors=covariates, timeline=np.linspace(0, 2))
fitter.print_summary(2)
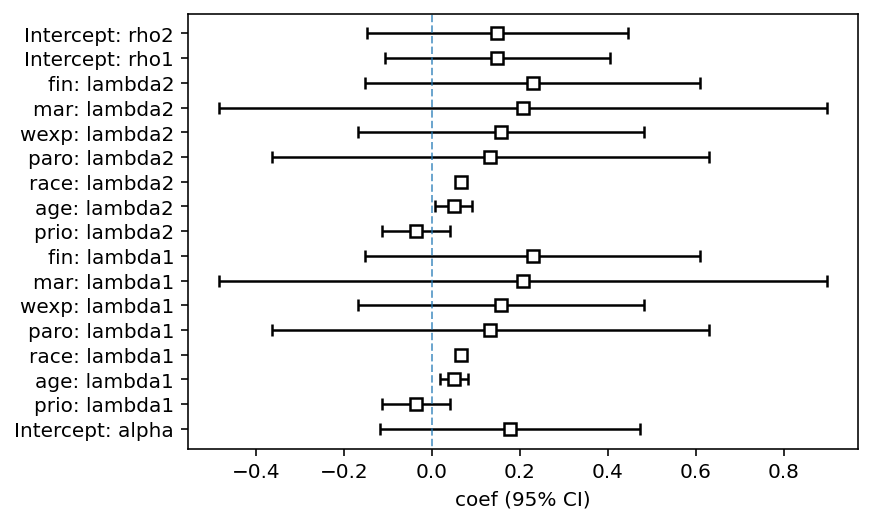
ax = fitter.plot()
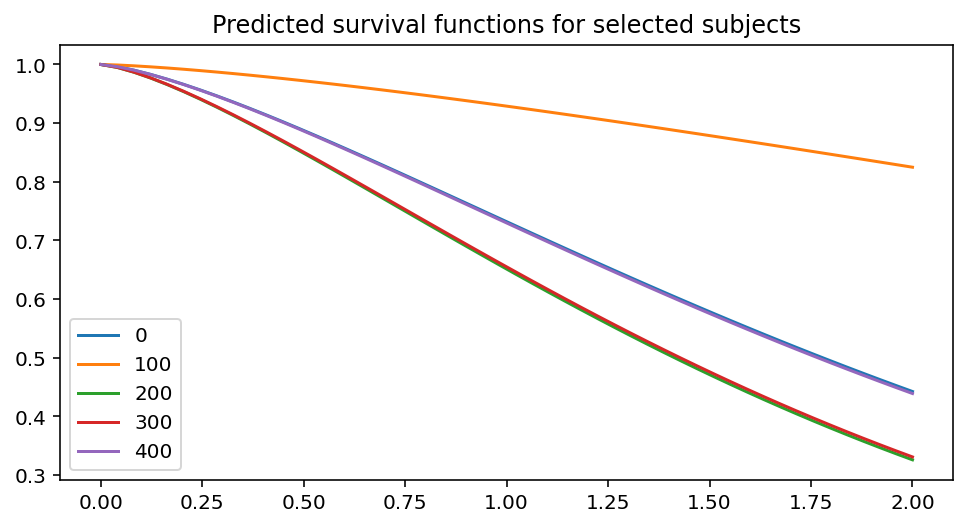
ax = fitter.predict_survival_function(rossi.loc[::100]).plot(figsize=(8, 4))
ax.set_title("Predicted survival functions for selected subjects")
/Users/camerondavidson-pilon/code/lifelines/lifelines/fitters/__init__.py:1985: StatisticalWarning: The diagonal of the variance_matrix_ has negative values. This could be a problem with DependentCompetingRisksHazard's fit to the data.
It's advisable to not trust the variances reported, and to be suspicious of the fitted parameters too.
warnings.warn(warning_text, exceptions.StatisticalWarning)
| model | lifelines.DependentCompetingRisksHazard |
|---|---|
| duration col | 'week' |
| event col | 'arrest' |
| penalizer | 0.1 |
| number of observations | 432 |
| number of events observed | 114 |
| log-likelihood | -239.19 |
| time fit was run | 2020-07-26 22:06:55 UTC |
| coef | exp(coef) | se(coef) | coef lower 95% | coef upper 95% | exp(coef) lower 95% | exp(coef) upper 95% | z | p | -log2(p) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| param | covariate | ||||||||||
| lambda1 | age | 0.05 | 1.05 | 0.02 | 0.02 | 0.08 | 1.02 | 1.09 | 3.06 | <0.005 | 8.80 |
| fin | 0.23 | 1.26 | 0.19 | -0.15 | 0.61 | 0.86 | 1.84 | 1.18 | 0.24 | 2.06 | |
| mar | 0.21 | 1.23 | 0.35 | -0.48 | 0.90 | 0.62 | 2.46 | 0.59 | 0.56 | 0.84 | |
| paro | 0.13 | 1.14 | 0.25 | -0.36 | 0.63 | 0.69 | 1.88 | 0.52 | 0.60 | 0.74 | |
| prio | -0.04 | 0.96 | 0.04 | -0.11 | 0.04 | 0.89 | 1.04 | -0.93 | 0.35 | 1.50 | |
| race | 0.07 | 1.07 | nan | nan | nan | nan | nan | nan | nan | nan | |
| wexp | 0.16 | 1.17 | 0.17 | -0.17 | 0.48 | 0.85 | 1.62 | 0.94 | 0.35 | 1.53 | |
| lambda2 | age | 0.05 | 1.05 | 0.02 | 0.01 | 0.09 | 1.01 | 1.10 | 2.34 | 0.02 | 5.70 |
| fin | 0.23 | 1.26 | 0.19 | -0.15 | 0.61 | 0.86 | 1.84 | 1.18 | 0.24 | 2.06 | |
| mar | 0.21 | 1.23 | 0.35 | -0.48 | 0.90 | 0.62 | 2.46 | 0.59 | 0.56 | 0.84 | |
| paro | 0.13 | 1.14 | 0.25 | -0.36 | 0.63 | 0.69 | 1.88 | 0.52 | 0.60 | 0.74 | |
| prio | -0.04 | 0.96 | 0.04 | -0.11 | 0.04 | 0.89 | 1.04 | -0.93 | 0.35 | 1.50 | |
| race | 0.07 | 1.07 | nan | nan | nan | nan | nan | nan | nan | nan | |
| wexp | 0.16 | 1.17 | 0.17 | -0.17 | 0.48 | 0.85 | 1.62 | 0.94 | 0.35 | 1.53 | |
| rho1 | Intercept | 0.15 | 1.16 | 0.13 | -0.11 | 0.40 | 0.90 | 1.50 | 1.14 | 0.25 | 1.98 |
| rho2 | Intercept | 0.15 | 1.16 | 0.15 | -0.15 | 0.44 | 0.86 | 1.56 | 0.99 | 0.32 | 1.62 |
| alpha | Intercept | 0.18 | 1.19 | 0.15 | -0.12 | 0.47 | 0.89 | 1.61 | 1.18 | 0.24 | 2.06 |
| AIC | 512.38 |
|---|---|
| log-likelihood ratio test | 127.04 on 12 df |
| -log2(p) of ll-ratio test | 68.48 |
[2]:
Text(0.5, 1.0, 'Predicted survival functions for selected subjects')
治愈模型¶
假设在我们的总体中有一个子总体永远不会经历感兴趣的事件。或者,对于某些个体来说,事件将在未来非常遥远的时刻发生,几乎等同于无限时间。在这种情况下,个体的生存函数不应渐近地趋近于零,而应趋近于某个正值。描述这种情况的模型有时被称为治愈模型(即个体“治愈”了死亡,因此不再易感)或时间滞后转换模型。
能够使用常见的生存模型并且包含一些“治愈”成分将会很好。假设对于那些会经历感兴趣事件的个体,他们的生存分布是威布尔分布,表示为\(S_W(t)\)。对于随机选择的个体,他们的生存曲线\(S(t)\)是:
\[\begin{split}\begin{align*}
S(t) = P(T > t) &= P(\text{cured}) P(T > t\;|\;\text{cured}) + P(\text{not cured}) P(T > t\;|\;\text{not cured}) \\
&= p + (1-p) S_W(t)
\end{align*}\end{split}\]
尽管它以一种非常规的形式存在,我们仍然可以确定累积风险(这是生存函数的负对数):
\[H(t) = -\log{\left(p + (1-p) S_W(t)\right)}\]
[3]:
from autograd.scipy.special import expit
class CureModel(ParametricRegressionFitter):
_scipy_fit_method = "SLSQP"
_scipy_fit_options = {"ftol": 1e-10, "maxiter": 200}
_fitted_parameter_names = ["lambda_", "beta_", "rho_"]
def _cumulative_hazard(self, params, T, Xs):
c = expit(np.dot(Xs["beta_"], params["beta_"]))
lambda_ = np.exp(np.dot(Xs["lambda_"], params["lambda_"]))
rho_ = np.exp(np.dot(Xs["rho_"], params["rho_"]))
sf = np.exp(-(T / lambda_) ** rho_)
return -np.log((1 - c) + c * sf)
cm = CureModel(penalizer=0.0)
rossi = load_rossi()
covariates = {"lambda_": rossi.columns.difference(['week', 'arrest']), "rho_": "1", "beta_": 'fin + 1'}
cm.fit(rossi, "week", event_col="arrest", regressors=covariates, timeline=np.arange(250))
cm.print_summary(2)
| model | lifelines.CureModel |
|---|---|
| duration col | 'week' |
| event col | 'arrest' |
| number of observations | 432 |
| number of events observed | 114 |
| log-likelihood | -702.64 |
| time fit was run | 2020-07-26 22:07:13 UTC |
| coef | exp(coef) | se(coef) | coef lower 95% | coef upper 95% | exp(coef) lower 95% | exp(coef) upper 95% | z | p | -log2(p) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| param | covariate | ||||||||||
| lambda_ | age | 0.16 | 1.17 | 0.05 | 0.05 | 0.26 | 1.06 | 1.30 | 2.96 | <0.005 | 8.36 |
| fin | 1.14 | 3.13 | 4320.37 | -8466.63 | 8468.92 | 0.00 | inf | 0.00 | 1.00 | 0.00 | |
| mar | 0.35 | 1.42 | 0.32 | -0.27 | 0.97 | 0.76 | 2.64 | 1.11 | 0.27 | 1.90 | |
| paro | 0.26 | 1.30 | 0.24 | -0.20 | 0.72 | 0.82 | 2.06 | 1.10 | 0.27 | 1.88 | |
| prio | -0.02 | 0.98 | 0.04 | -0.10 | 0.06 | 0.91 | 1.06 | -0.51 | 0.61 | 0.71 | |
| race | 0.23 | 1.26 | 0.28 | -0.32 | 0.79 | 0.72 | 2.20 | 0.82 | 0.41 | 1.29 | |
| wexp | 0.25 | 1.28 | 0.17 | -0.08 | 0.57 | 0.92 | 1.78 | 1.49 | 0.14 | 2.86 | |
| rho_ | Intercept | 0.13 | 1.14 | 0.08 | -0.03 | 0.29 | 0.97 | 1.34 | 1.57 | 0.12 | 3.11 |
| beta_ | Intercept | 0.18 | 1.19 | 0.10 | -0.02 | 0.37 | 0.98 | 1.45 | 1.75 | 0.08 | 3.64 |
| fin | 15.80 | 7.30e+06 | 0.17 | 15.46 | 16.14 | 5.20e+06 | 1.03e+07 | 90.99 | <0.005 | inf |
| AIC | 1425.29 |
|---|---|
| log-likelihood ratio test | -12.04 on 7 df |
| -log2(p) of ll-ratio test | -0.00 |
[4]:
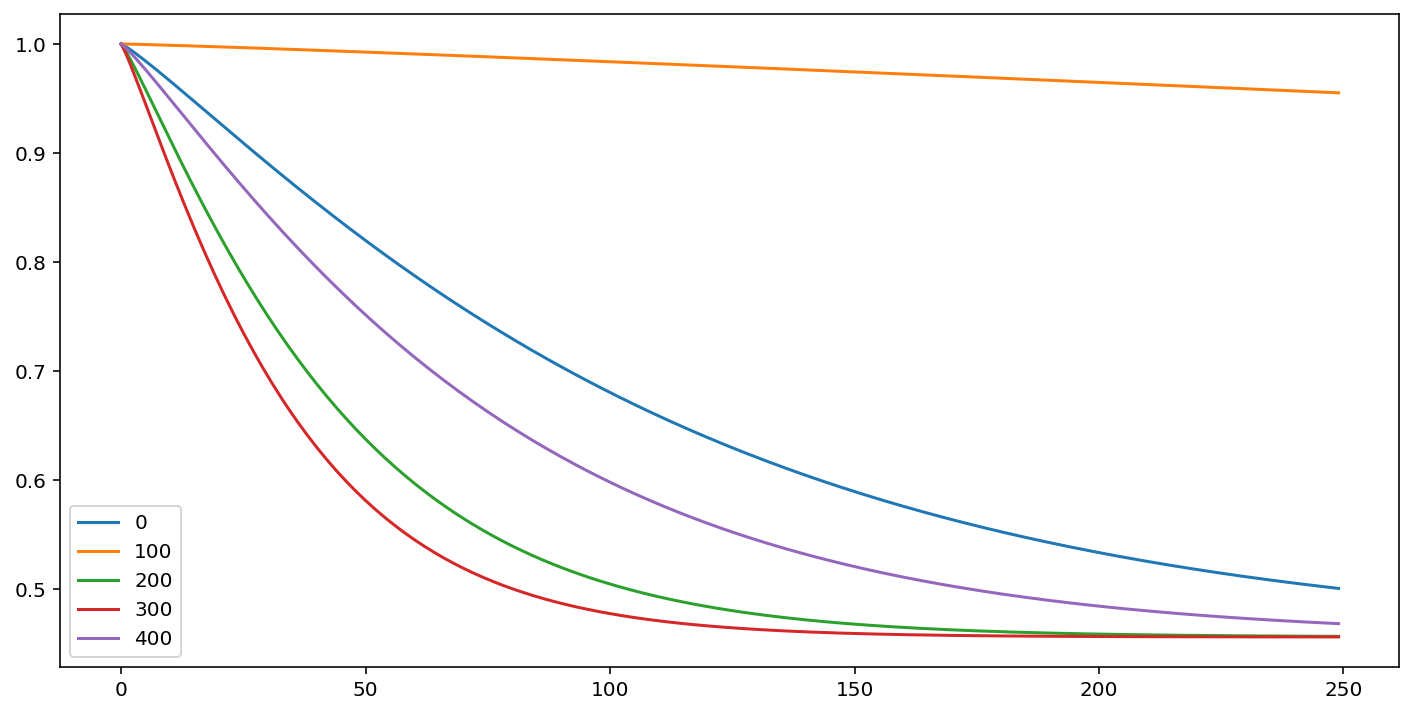
cm.predict_survival_function(rossi.loc[::100]).plot(figsize=(12,6))
[4]:
<AxesSubplot:>
[5]:
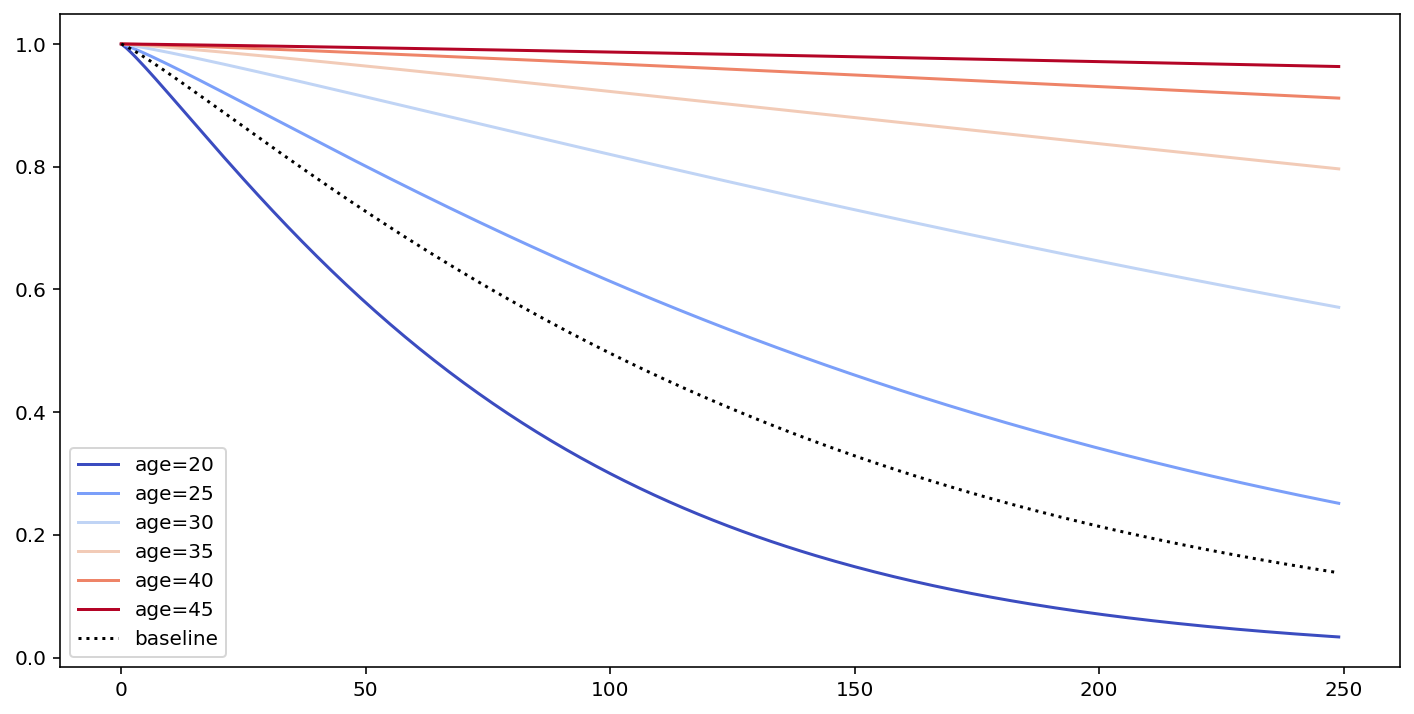
# what's the effect on the survival curve if I vary "age"
fig, ax = plt.subplots(figsize=(12, 6))
cm.plot_covariate_groups(['age'], values=np.arange(20, 50, 5), cmap='coolwarm', ax=ax)
[5]:
<AxesSubplot:>
样条模型¶
请查看示例文件夹中的 royston_parmar_splines.py 文件:https://github.com/CamDavidsonPilon/lifelines/tree/master/examples