因果树/森林处理效果估计与树可视化
[1]:
%reload_ext autoreload
%autoreload 2
%matplotlib inline
[2]:
import pandas as pd
import numpy as np
import multiprocessing as mp
from collections import defaultdict
np.random.seed(42)
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
import causalml
from causalml.metrics import plot_gain, plot_qini, qini_score
from causalml.dataset import synthetic_data
from causalml.inference.tree import plot_dist_tree_leaves_values, get_tree_leaves_mask
from causalml.inference.meta import BaseSRegressor, BaseXRegressor, BaseTRegressor, BaseDRRegressor
from causalml.inference.tree import CausalRandomForestRegressor
from causalml.inference.tree import CausalTreeRegressor
from causalml.inference.tree.plot import plot_causal_tree
import matplotlib.pyplot as plt
import seaborn as sns
%config InlineBackend.figure_format = 'retina'
Failed to import duecredit due to No module named 'duecredit'
[3]:
import importlib
print(importlib.metadata.version('causalml') )
0.15.3.dev0
[4]:
# Simulate randomized trial: mode=2
y, X, w, tau, b, e = synthetic_data(mode=2, n=15000, p=20, sigma=5.5)
df = pd.DataFrame(X)
feature_names = [f'feature_{i}' for i in range(X.shape[1])]
df.columns = feature_names
df['outcome'] = y
df['treatment'] = w
df['treatment_effect'] = tau
[5]:
df.head()
[5]:
| 特征_0 | 特征_1 | 特征_2 | 特征_3 | 特征_4 | 特征_5 | 特征_6 | 特征_7 | 特征_8 | 特征_9 | ... | 特征_13 | 特征_14 | 特征_15 | 特征_16 | 特征_17 | 特征_18 | 特征_19 | 结果 | 处理 | 处理效果 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.496714 | -0.138264 | 0.647689 | 1.523030 | -0.234153 | -0.234137 | 1.579213 | 0.767435 | -0.469474 | 0.542560 | ... | -1.913280 | -1.724918 | -0.562288 | -1.012831 | 0.314247 | -0.908024 | -1.412304 | 7.413595 | 1 | 1.123117 |
| 1 | 1.465649 | -0.225776 | 0.067528 | -1.424748 | -0.544383 | 0.110923 | -1.150994 | 0.375698 | -0.600639 | -0.291694 | ... | -1.057711 | 0.822545 | -1.220844 | 0.208864 | -1.959670 | -1.328186 | 0.196861 | -11.263144 | 0 | 2.052266 |
| 2 | 0.738467 | 0.171368 | -0.115648 | -0.301104 | -1.478522 | -0.719844 | -0.460639 | 1.057122 | 0.343618 | -1.763040 | ... | 0.611676 | 1.031000 | 0.931280 | -0.839218 | -0.309212 | 0.331263 | 0.975545 | 0.269378 | 0 | 1.520964 |
| 3 | -0.479174 | -0.185659 | -1.106335 | -1.196207 | 0.812526 | 1.356240 | -0.072010 | 1.003533 | 0.361636 | -0.645120 | ... | 1.564644 | -2.619745 | 0.821903 | 0.087047 | -0.299007 | 0.091761 | -1.987569 | -0.976893 | 0 | 0.125446 |
| 4 | -0.219672 | 0.357113 | 1.477894 | -0.518270 | -0.808494 | -0.501757 | 0.915402 | 0.328751 | -0.529760 | 0.513267 | ... | -0.327662 | -0.392108 | -1.463515 | 0.296120 | 0.261055 | 0.005113 | -0.234587 | -0.608710 | 1 | 0.667889 |
5 行 × 23 列
[6]:
# Look at the conversion rate and sample size in each group
df.pivot_table(values='outcome',
index='treatment',
aggfunc=[np.mean, np.size],
margins=True)
[6]:
| 平均值 | 大小 | |
|---|---|---|
| 结果 | 结果 | |
| 治疗 | ||
| 0 | 0.994413 | 7502 |
| 1 | 1.802171 | 7498 |
| 全部 | 1.398184 | 15000 |
[7]:
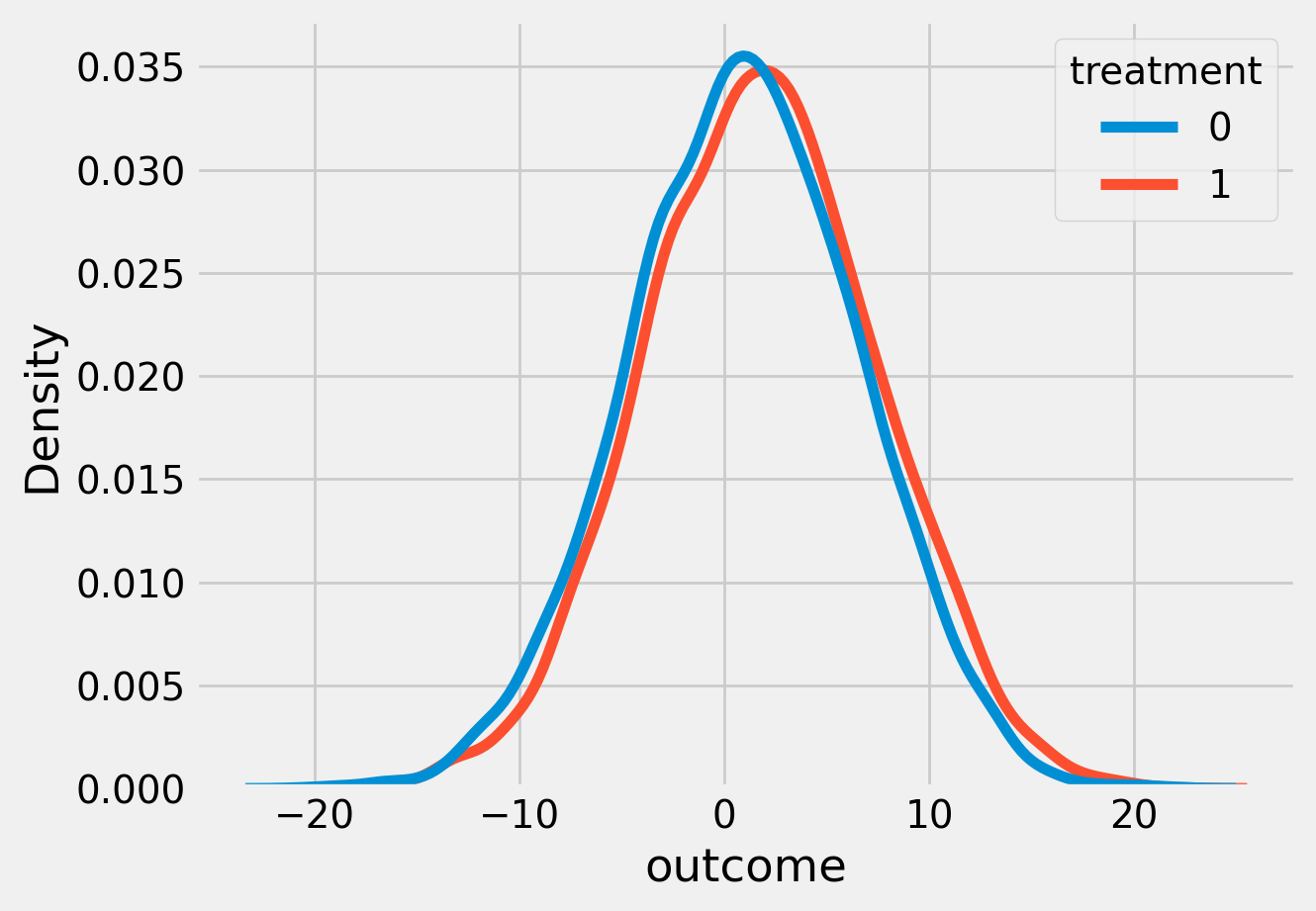
sns.kdeplot(data=df, x='outcome', hue='treatment')
plt.show()
[8]:
# Split data to training and testing samples for model validation (next section)
df_train, df_test = train_test_split(df, test_size=0.2, random_state=11101)
n_test = df_test.shape[0]
n_train = df_train.shape[0]
[9]:
# Table to gather estimated ITEs by models
df_result = pd.DataFrame({
'outcome': df_test['outcome'],
'is_treated': df_test['treatment'],
'treatment_effect': df_test['treatment_effect']
})
CausalTreeRegressor
因果树可用的标准:
standard_mse: scikit-learn 的均方误差,其中节点值存储 \(E_{node_i}(X|T=1)-E_{node_i}(X|T=0)\),即处理效果。
causal_mse: 该标准奖励那些能够发现治疗效果强烈异质性的分区,并惩罚那些在叶估计中产生方差的分区。 https://www.pnas.org/doi/10.1073/pnas.1510489113
[10]:
ctrees = {
'ctree_mse': {
'params':
dict(criterion='standard_mse',
control_name=0,
min_impurity_decrease=0,
min_samples_leaf=400,
groups_penalty=0.,
groups_cnt=True),
},
'ctree_cmse': {
'params':
dict(
criterion='causal_mse',
control_name=0,
min_samples_leaf=400,
groups_penalty=0.,
groups_cnt=True,
),
},
'ctree_cmse_p=0.1': {
'params':
dict(
criterion='causal_mse',
control_name=0,
min_samples_leaf=400,
groups_penalty=0.1,
groups_cnt=True,
),
},
'ctree_cmse_p=0.25': {
'params':
dict(
criterion='causal_mse',
control_name=0,
min_samples_leaf=400,
groups_penalty=0.25,
groups_cnt=True,
),
},
'ctree_cmse_p=0.5': {
'params':
dict(
criterion='causal_mse',
control_name=0,
min_samples_leaf=400,
groups_penalty=0.5,
groups_cnt=True,
),
},
'ctree_ttest': {
'params':
dict(criterion='t_test',
control_name=0,
min_samples_leaf=400,
groups_penalty=0.,
groups_cnt=True),
},
}
[11]:
# Model treatment effect
for ctree_name, ctree_info in ctrees.items():
print(f"Fitting: {ctree_name}")
ctree = CausalTreeRegressor(**ctree_info['params'])
ctree.fit(X=df_train[feature_names].values,
treatment=df_train['treatment'].values,
y=df_train['outcome'].values)
ctrees[ctree_name].update({'model': ctree})
df_result[ctree_name] = ctree.predict(df_test[feature_names].values)
Fitting: ctree_mse
Fitting: ctree_cmse
Fitting: ctree_cmse_p=0.1
Fitting: ctree_cmse_p=0.25
Fitting: ctree_cmse_p=0.5
Fitting: ctree_ttest
[12]:
df_result.head()
[12]:
| 结果 | 是否处理 | 处理效果 | ctree_mse | ctree_cmse | ctree_cmse_p=0.1 | ctree_cmse_p=0.25 | ctree_cmse_p=0.5 | ctree_ttest | |
|---|---|---|---|---|---|---|---|---|---|
| 625 | 3.519424 | 1 | 0.819201 | 1.624443 | -1.690532 | 0.129960 | -0.947096 | -0.947096 | -1.690532 |
| 5717 | -0.456031 | 0 | 1.131599 | 0.809237 | 0.367659 | 0.992395 | 1.978697 | 1.978697 | 1.054970 |
| 14801 | 4.479222 | 0 | 1.969727 | 1.624443 | -0.778434 | 3.388318 | 1.937710 | 1.937710 | 1.744661 |
| 13605 | 4.523891 | 0 | 0.884079 | 0.809237 | 0.367659 | 0.992395 | 0.805110 | 0.805110 | 1.039292 |
| 4208 | -4.615111 | 0 | 1.179124 | 0.809237 | 2.134070 | 0.992395 | 0.928345 | 0.928345 | 1.054970 |
[13]:
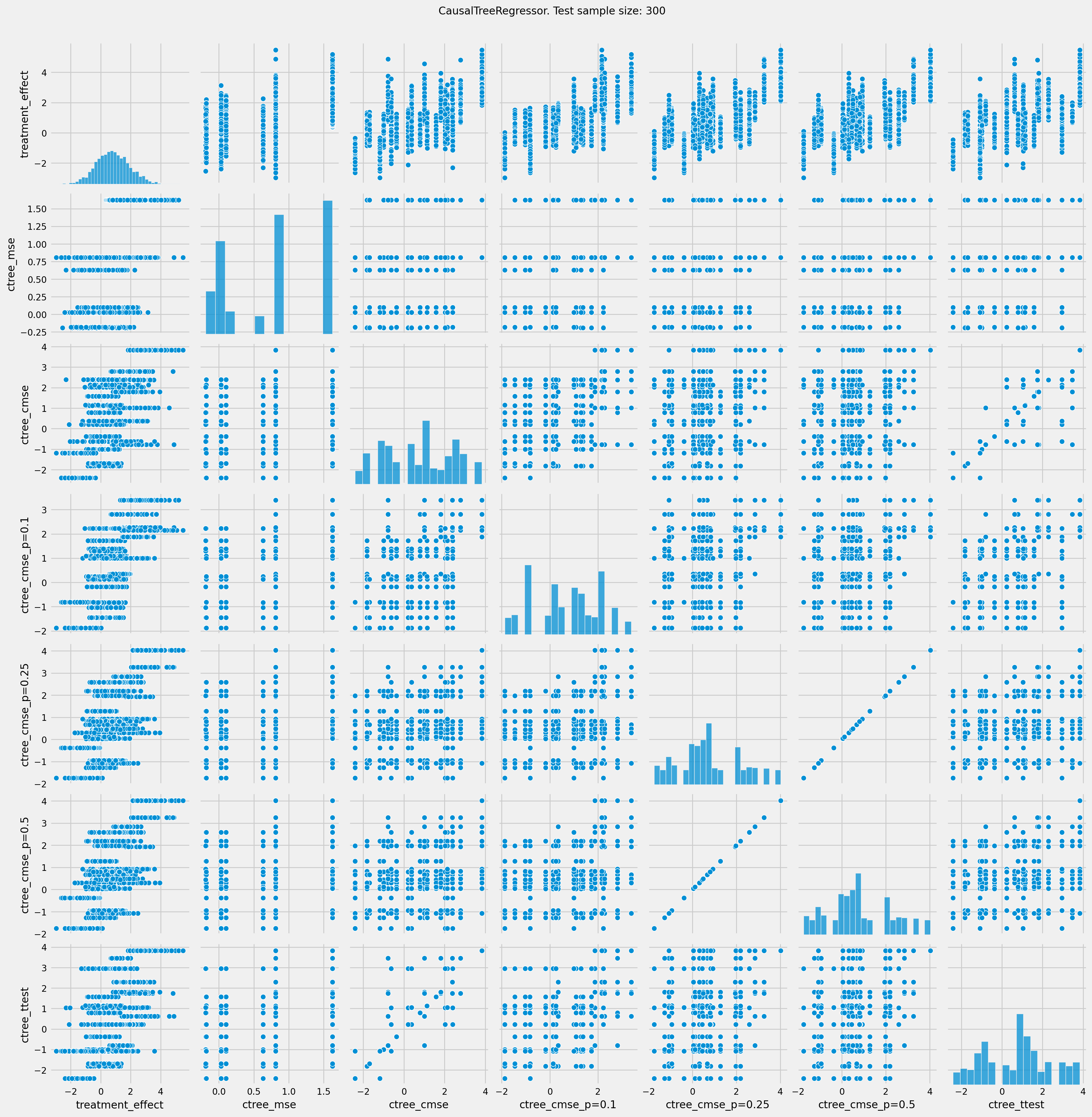
# See treatment effect estimation with CausalTreeRegressor vs true treatment effect
n_obs = 300
indxs = df_result.index.values
np.random.shuffle(indxs)
indxs = indxs[:n_obs]
plt.rcParams.update({'font.size': 10})
pairplot = sns.pairplot(df_result[['treatment_effect', *list(ctrees)]])
pairplot.fig.suptitle(f"CausalTreeRegressor. Test sample size: {n_obs}" , y=1.02)
plt.show()
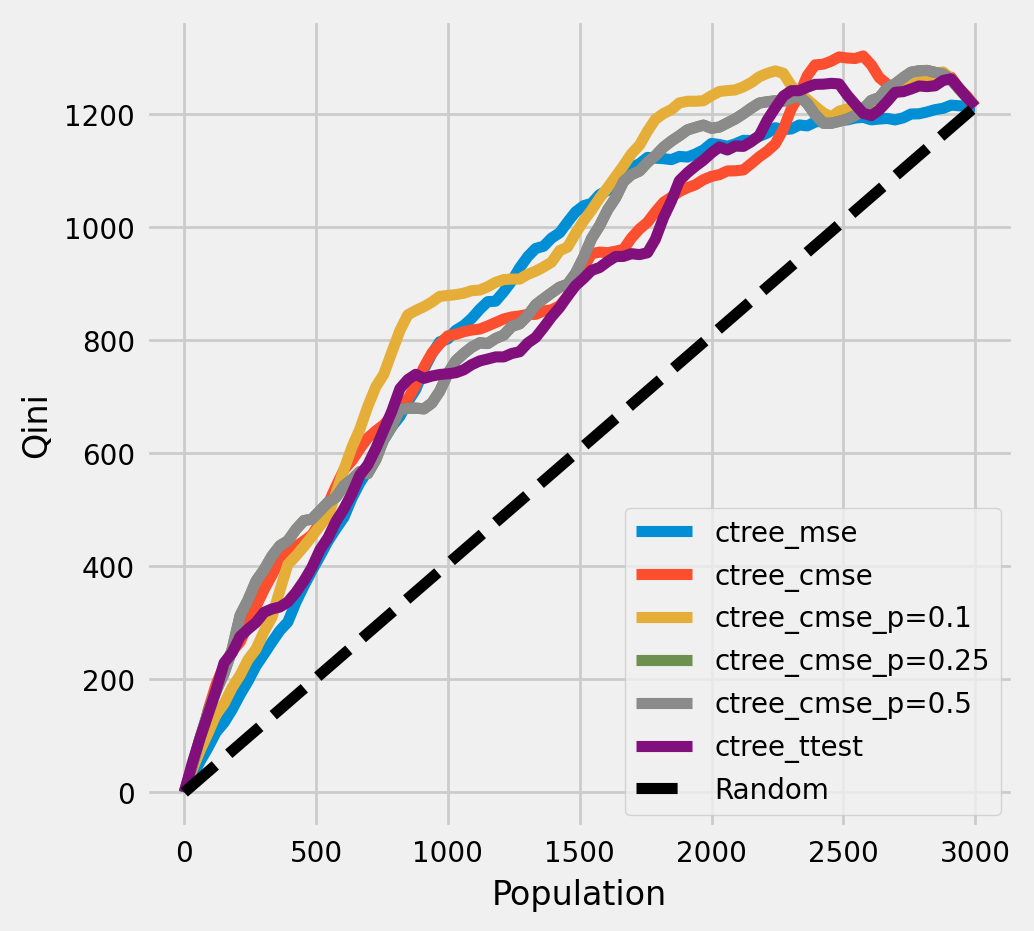
绘制Qini图表
[14]:
plot_qini(df_result,
outcome_col='outcome',
treatment_col='is_treated',
treatment_effect_col='treatment_effect',
figsize=(5,5)
)
[15]:
df_qini = qini_score(df_result,
outcome_col='outcome',
treatment_col='is_treated',
treatment_effect_col='treatment_effect')
df_qini.sort_values(ascending=False)
[15]:
ctree_cmse_p=0.1 0.256107
ctree_cmse_p=0.25 0.228042
ctree_cmse_p=0.5 0.228042
ctree_cmse 0.218939
ctree_mse 0.214093
ctree_ttest 0.201714
dtype: float64
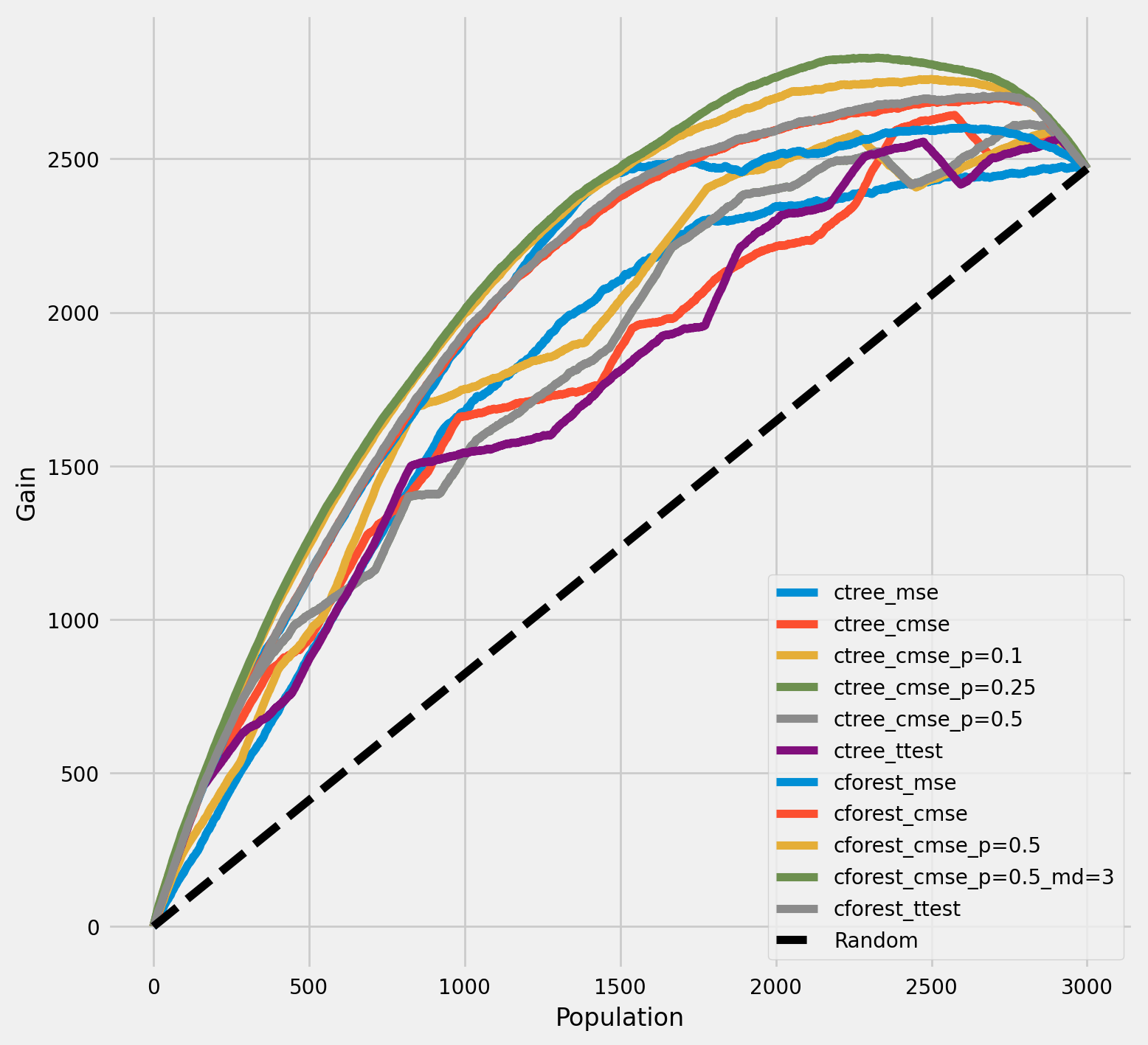
每个群体中真实治疗效果的累积增益
[16]:
plot_gain(df_result,
outcome_col='outcome',
treatment_col='is_treated',
treatment_effect_col='treatment_effect',
n = n_test,
figsize=(5,5)
)
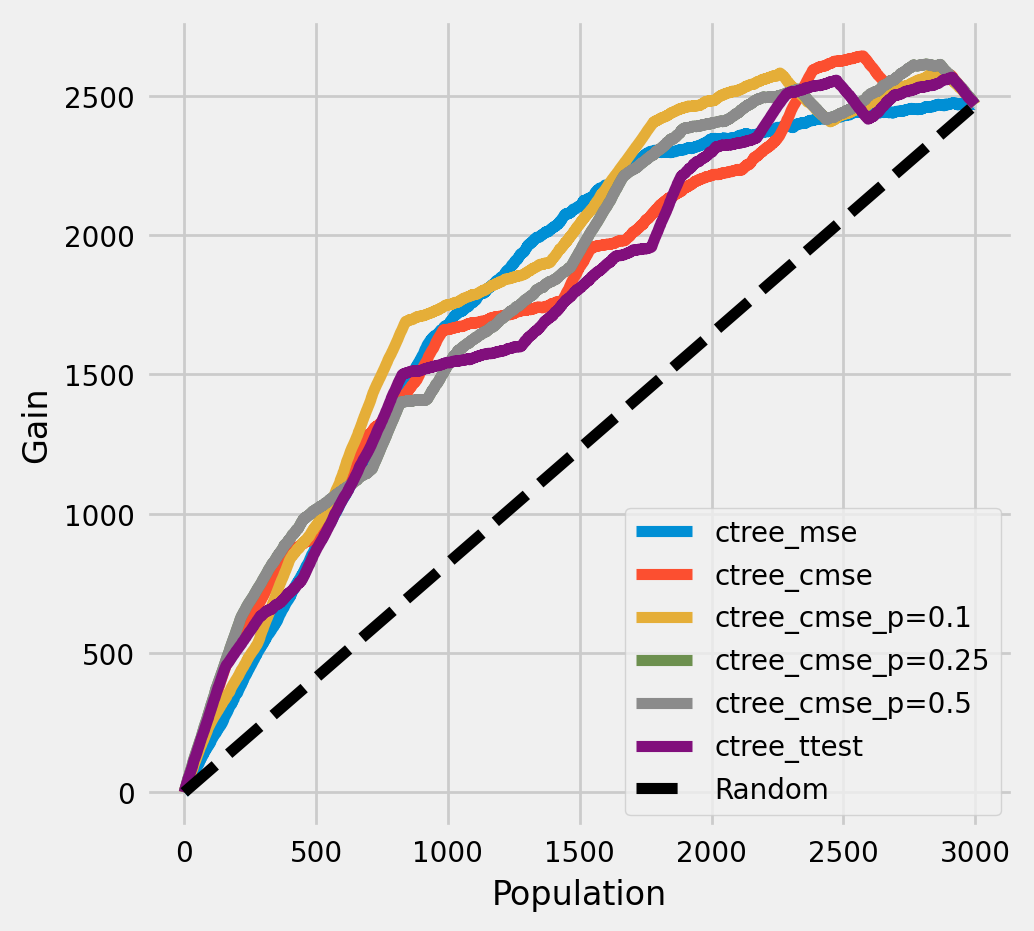
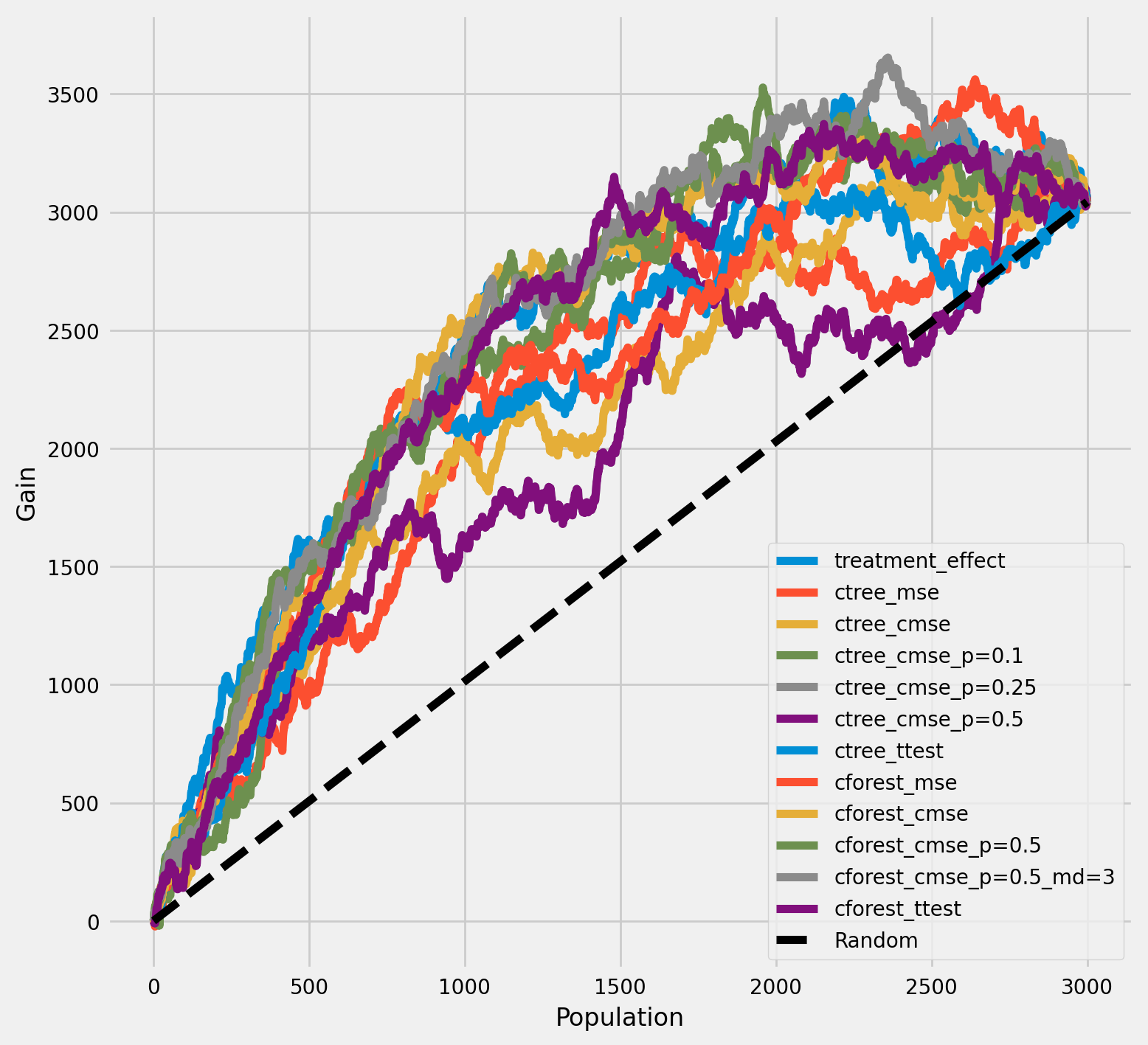
每个群体中治疗组和对照组平均结果之间的累积差异
[17]:
plot_gain(df_result,
outcome_col='outcome',
treatment_col='is_treated',
n = n_test,
figsize=(5,5)
)
使用sklearn函数绘制树并保存为矢量图形
[18]:
for ctree_name, ctree_info in ctrees.items():
plt.figure(figsize=(20,20))
plot_causal_tree(ctree_info['model'],
feature_names = feature_names,
filled=True,
impurity=True,
proportion=False,
)
plt.title(ctree_name)
plt.savefig(f'{ctree_name}.svg')
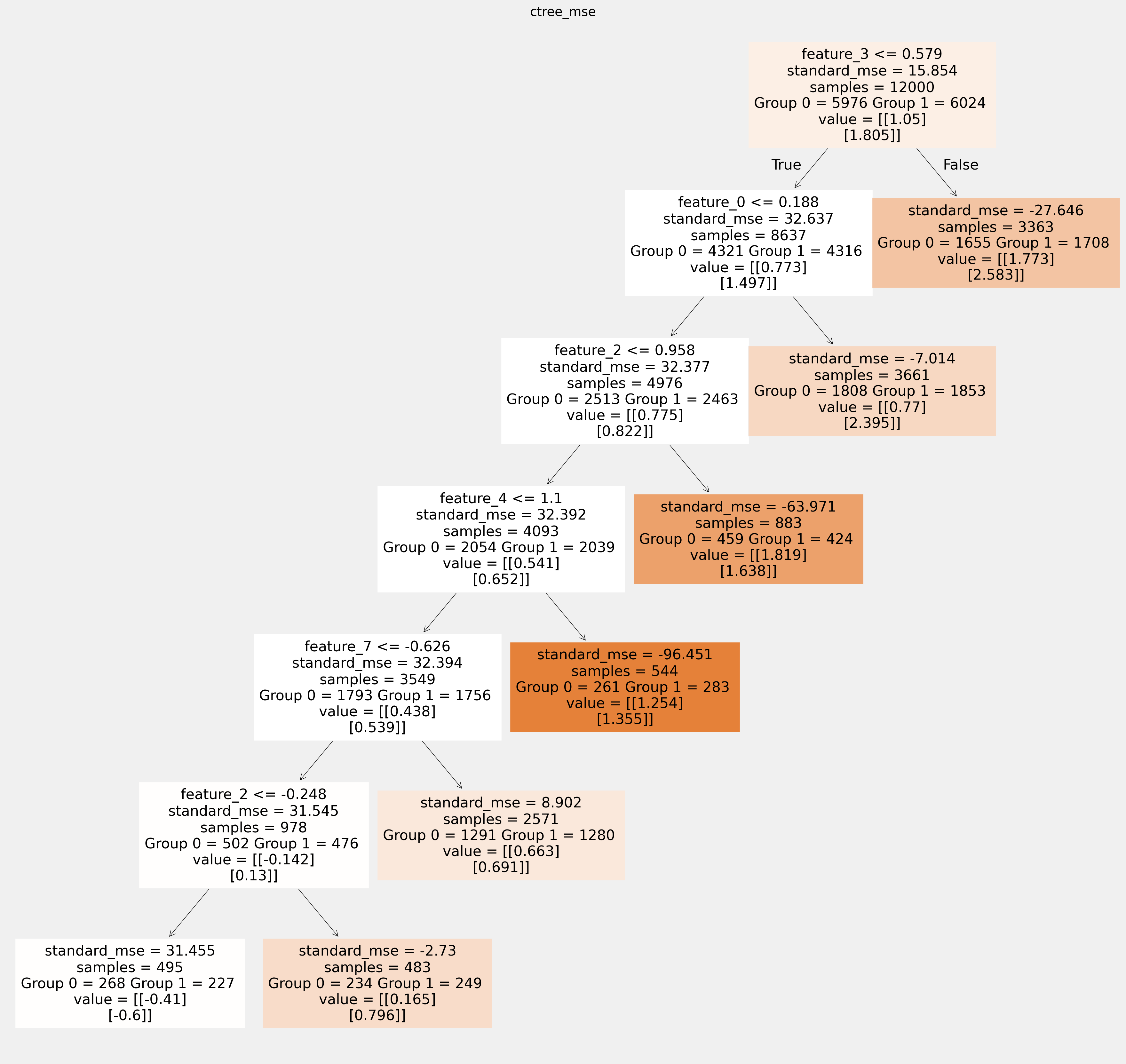
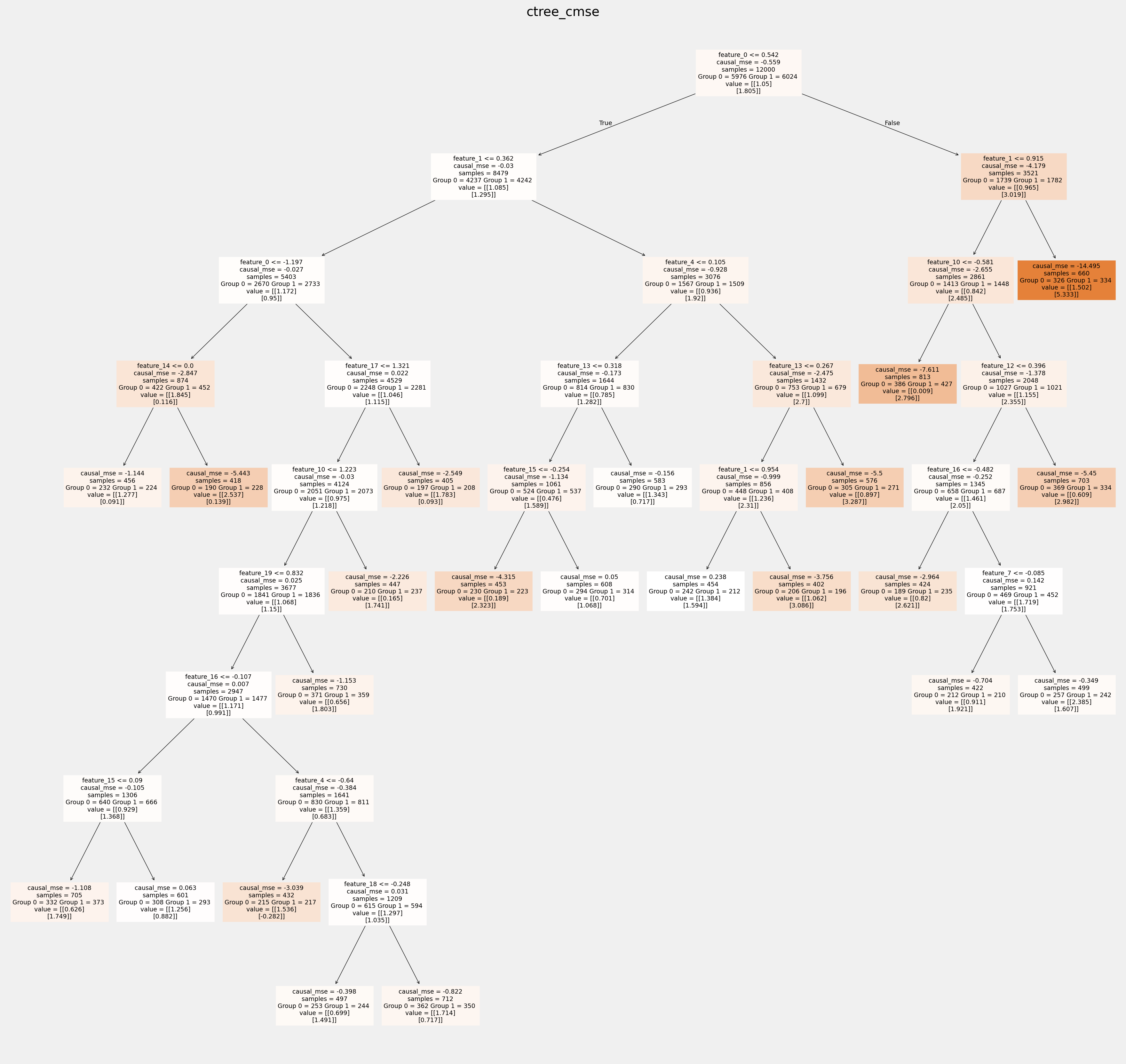
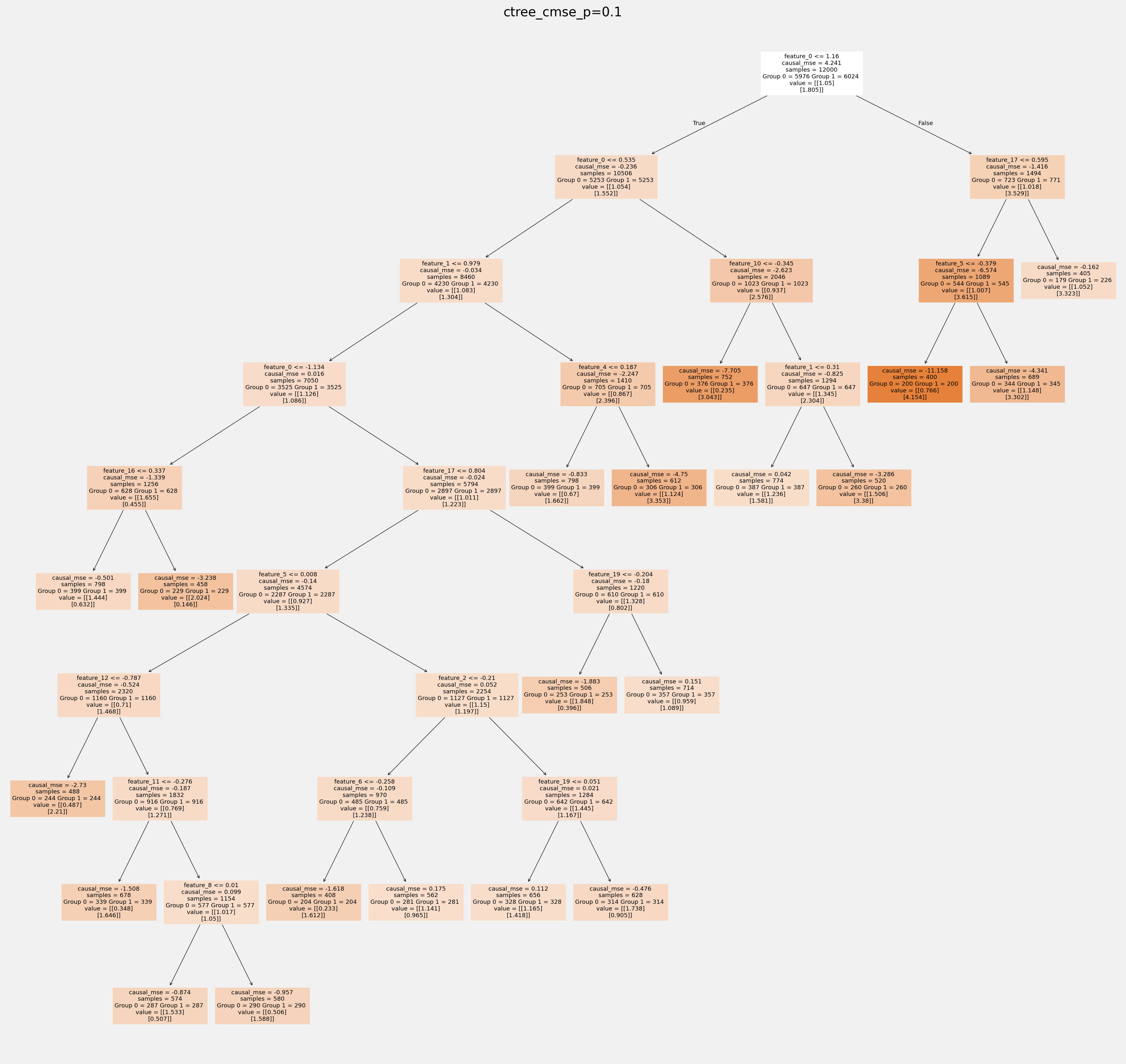

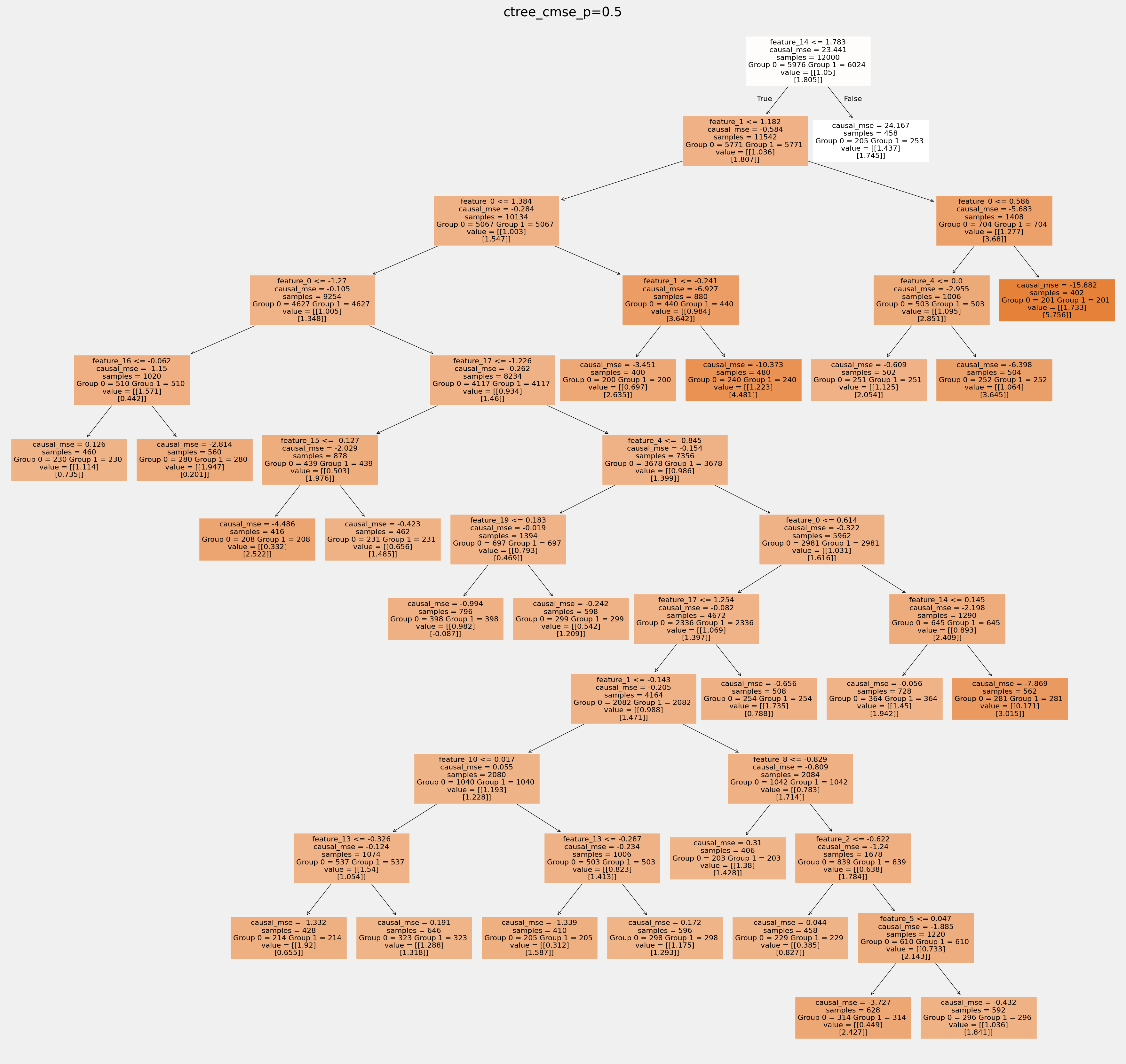

拟合树中叶节点的值如何相互不同:
[19]:
for ctree_name, ctree_info in ctrees.items():
plot_dist_tree_leaves_values(ctree_info['model'],
figsize=(3,3),
title=f'Tree({ctree_name}) leaves values distribution')


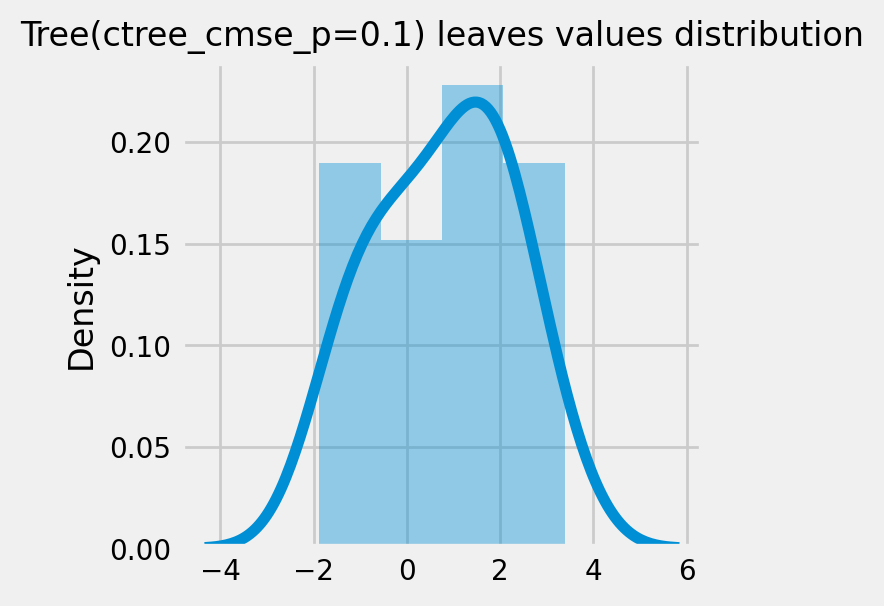
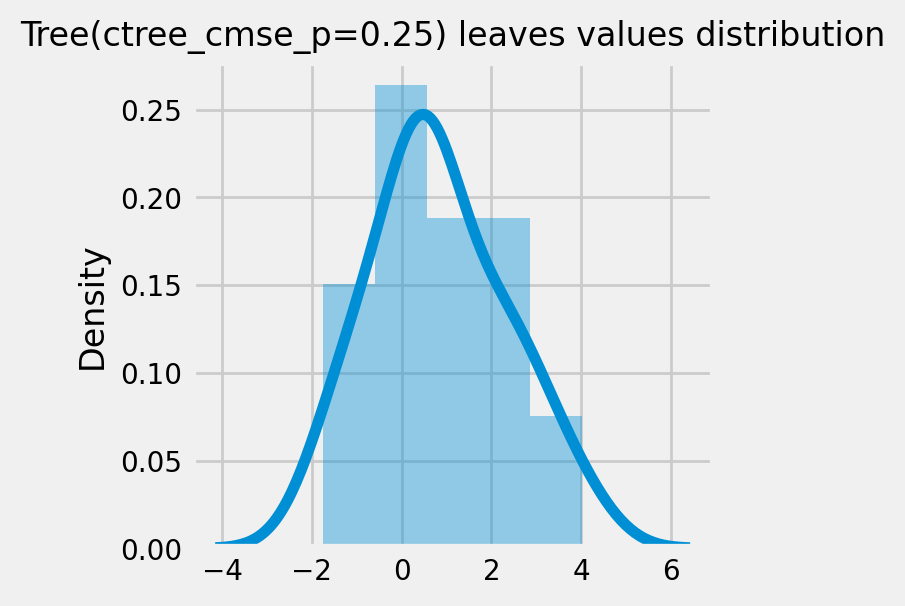
CausalRandomForestRegressor
[20]:
cforests = {
'cforest_mse': {
'params':
dict(criterion='standard_mse',
control_name=0,
min_impurity_decrease=0,
min_samples_leaf=400,
groups_penalty=0.,
groups_cnt=True),
},
'cforest_cmse': {
'params':
dict(
criterion='causal_mse',
control_name=0,
min_samples_leaf=400,
groups_penalty=0.,
groups_cnt=True
),
},
'cforest_cmse_p=0.5': {
'params':
dict(
criterion='causal_mse',
control_name=0,
min_samples_leaf=400,
groups_penalty=0.5,
groups_cnt=True,
),
},
'cforest_cmse_p=0.5_md=3': {
'params':
dict(
criterion='causal_mse',
control_name=0,
max_depth=3,
min_samples_leaf=400,
groups_penalty=0.5,
groups_cnt=True,
),
},
'cforest_ttest': {
'params':
dict(criterion='t_test',
control_name=0,
min_samples_leaf=400,
groups_penalty=0.,
groups_cnt=True),
},
}
[21]:
# Model treatment effect
for cforest_name, cforest_info in cforests.items():
print(f"Fitting: {cforest_name}")
cforest = CausalRandomForestRegressor(**cforest_info['params'])
cforest.fit(X=df_train[feature_names].values,
treatment=df_train['treatment'].values,
y=df_train['outcome'].values)
cforests[cforest_name].update({'model': cforest})
df_result[cforest_name] = cforest.predict(df_test[feature_names].values)
Fitting: cforest_mse
Fitting: cforest_cmse
Fitting: cforest_cmse_p=0.5
Fitting: cforest_cmse_p=0.5_md=3
Fitting: cforest_ttest
[22]:
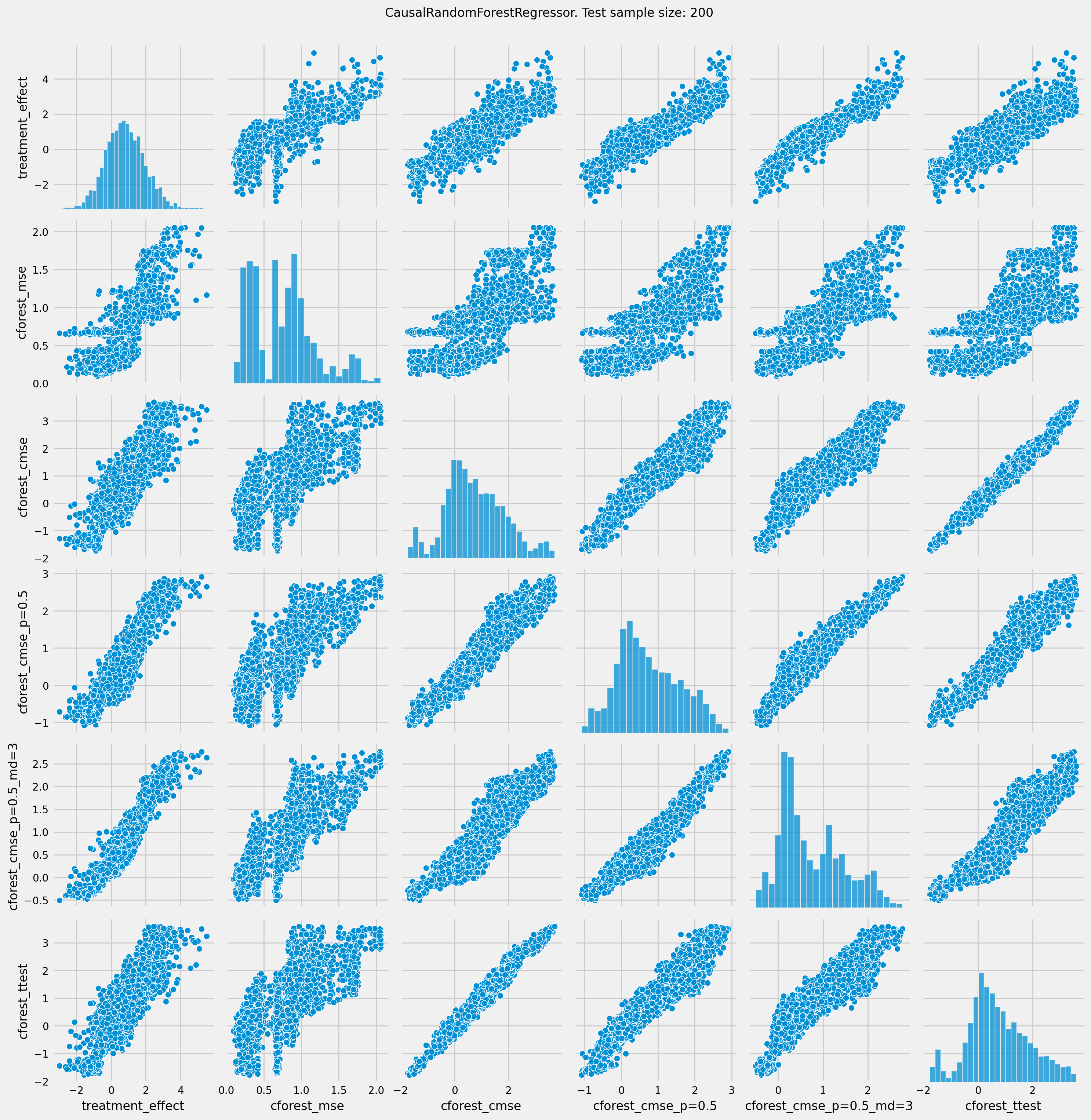
# See treatment effect estimation with CausalRandomForestRegressor vs true treatment effect
n_obs = 200
indxs = df_result.index.values
np.random.shuffle(indxs)
indxs = indxs[:n_obs]
plt.rcParams.update({'font.size': 10})
pairplot = sns.pairplot(df_result[['treatment_effect', *list(cforests)]])
pairplot.fig.suptitle(f"CausalRandomForestRegressor. Test sample size: {n_obs}" , y=1.02)
plt.show()
[23]:
df_qini = qini_score(df_result,
outcome_col='outcome',
treatment_col='is_treated',
treatment_effect_col='treatment_effect')
df_qini.sort_values(ascending=False)
[23]:
cforest_cmse_p=0.5_md=3 0.368213
cforest_cmse_p=0.5 0.351258
cforest_ttest 0.326421
cforest_cmse 0.324050
cforest_mse 0.308017
ctree_cmse_p=0.1 0.256107
ctree_cmse_p=0.25 0.228042
ctree_cmse_p=0.5 0.228042
ctree_cmse 0.218939
ctree_mse 0.214093
ctree_ttest 0.201714
dtype: float64
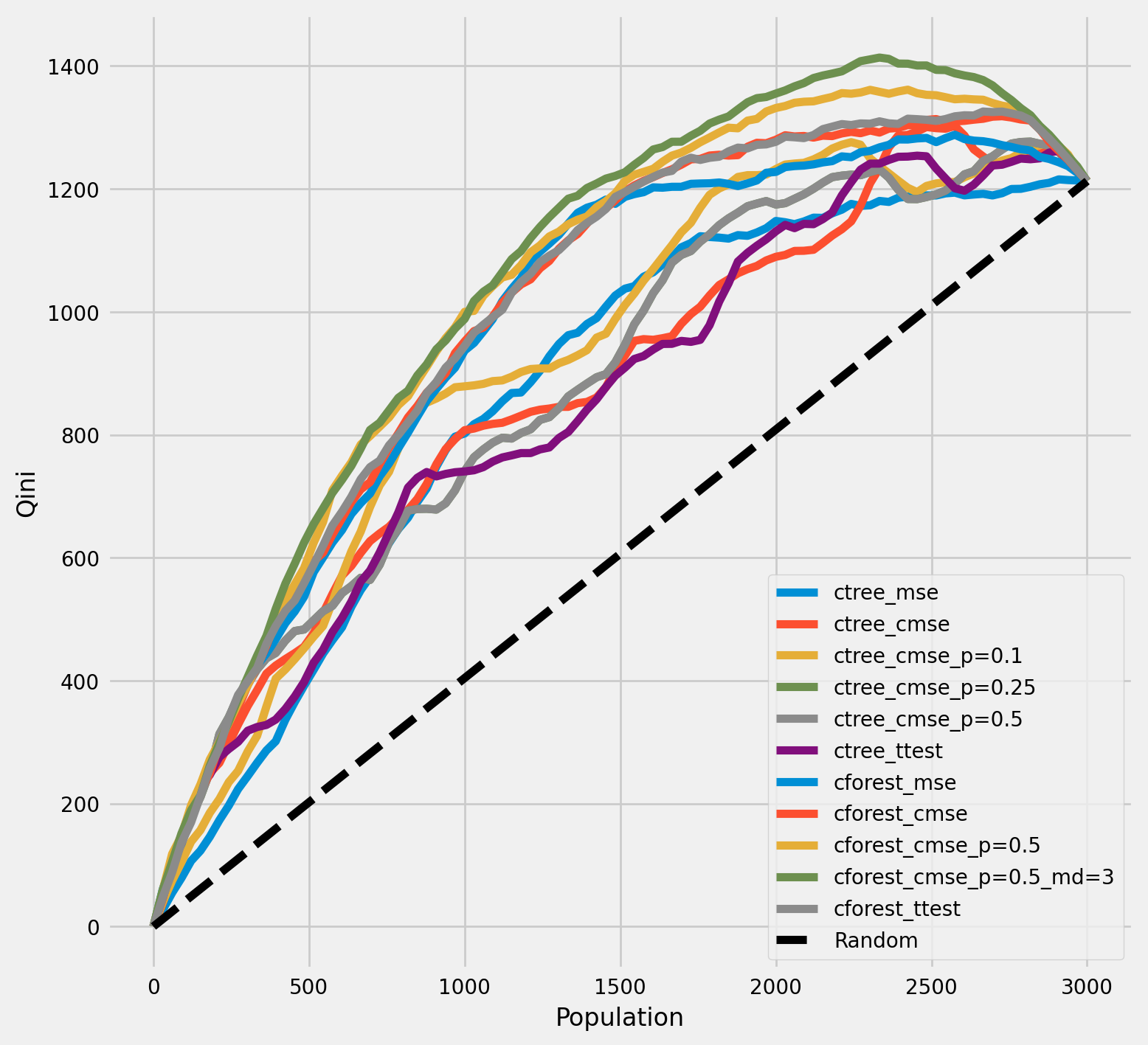
七牛图表
[24]:
plot_qini(df_result,
outcome_col='outcome',
treatment_col='is_treated',
treatment_effect_col='treatment_effect',
figsize=(8,8)
)
[25]:
df_qini = qini_score(df_result,
outcome_col='outcome',
treatment_col='is_treated',
treatment_effect_col='treatment_effect')
df_qini.sort_values(ascending=False)
[25]:
cforest_cmse_p=0.5_md=3 0.368213
cforest_cmse_p=0.5 0.351258
cforest_ttest 0.326421
cforest_cmse 0.324050
cforest_mse 0.308017
ctree_cmse_p=0.1 0.256107
ctree_cmse_p=0.25 0.228042
ctree_cmse_p=0.5 0.228042
ctree_cmse 0.218939
ctree_mse 0.214093
ctree_ttest 0.201714
dtype: float64
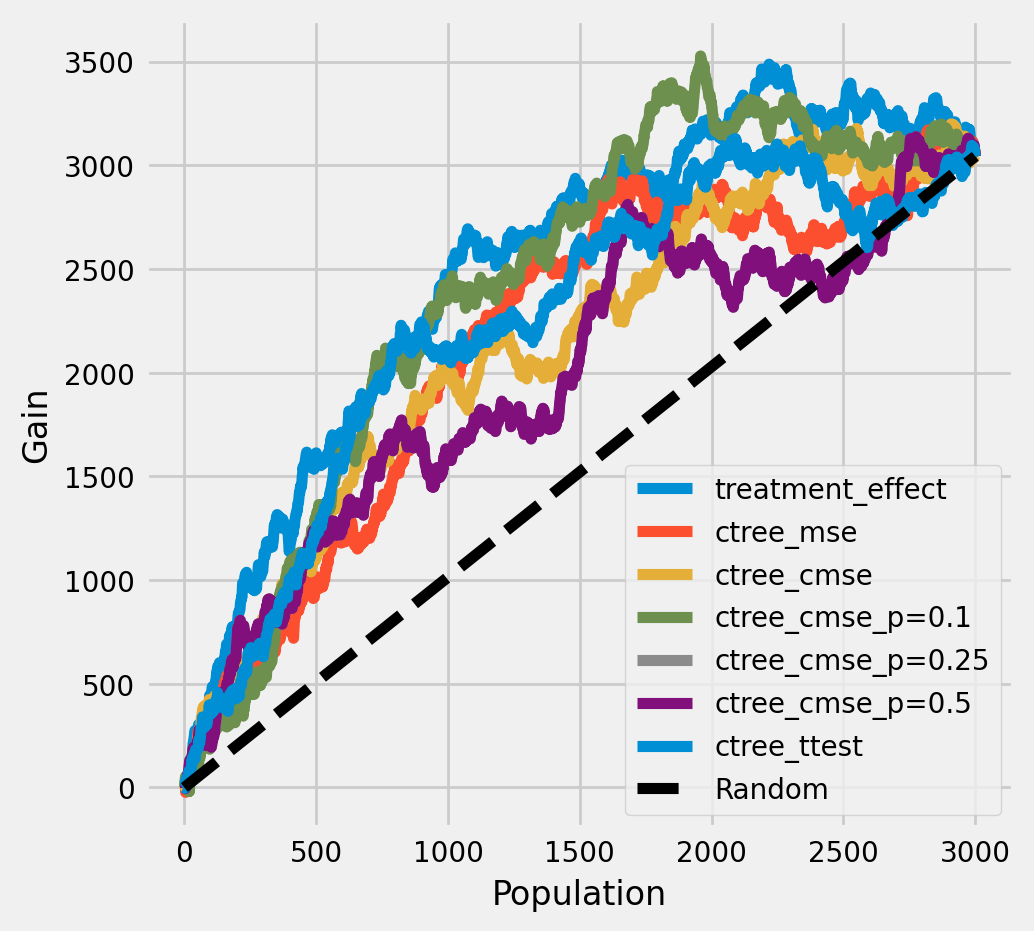
每个群体中真实治疗效果的累积增益
[26]:
plot_gain(df_result,
outcome_col='outcome',
treatment_col='is_treated',
treatment_effect_col='treatment_effect',
n = n_test
)
每个群体中治疗组和对照组平均结果之间的累积差异
[27]:
plot_gain(df_result,
outcome_col='outcome',
treatment_col='is_treated',
n = n_test
)
元学习算法
[28]:
X_train = df_train[feature_names].values
X_test = df_test[feature_names].values
# learner - DecisionTreeRegressor
# treatment learner - LinearRegression()
learner_x = BaseXRegressor(learner=DecisionTreeRegressor(),
treatment_effect_learner=LinearRegression())
learner_s = BaseSRegressor(learner=DecisionTreeRegressor())
learner_t = BaseTRegressor(learner=DecisionTreeRegressor(),
treatment_learner=LinearRegression())
learner_dr = BaseDRRegressor(learner=DecisionTreeRegressor(),
treatment_effect_learner=LinearRegression())
learner_x.fit(X=X_train, treatment=df_train['treatment'].values, y=df_train['outcome'].values)
learner_s.fit(X=X_train, treatment=df_train['treatment'].values, y=df_train['outcome'].values)
learner_t.fit(X=X_train, treatment=df_train['treatment'].values, y=df_train['outcome'].values)
learner_dr.fit(X=X_train, treatment=df_train['treatment'].values, y=df_train['outcome'].values)
df_result['learner_x_ite'] = learner_x.predict(X_test)
df_result['learner_s_ite'] = learner_s.predict(X_test)
df_result['learner_t_ite'] = learner_t.predict(X_test)
df_result['learner_dr_ite'] = learner_dr.predict(X_test)
[29]:
cate_dr = learner_dr.predict(X)
cate_x = learner_x.predict(X)
cate_s = learner_s.predict(X)
cate_t = learner_t.predict(X)
cate_ctrees = [info['model'].predict(X) for _, info in ctrees.items()]
cate_cforests = [info['model'].predict(X) for _, info in cforests.items()]
model_cate = [
*cate_ctrees,
*cate_cforests,
cate_x, cate_s, cate_t, cate_dr
]
model_names = [
*list(ctrees), *list(cforests),
'X Learner', 'S Learner', 'T Learner', 'DR Learner']
[30]:
plot_gain(df_result,
outcome_col='outcome',
treatment_col='is_treated',
n = n_test
)
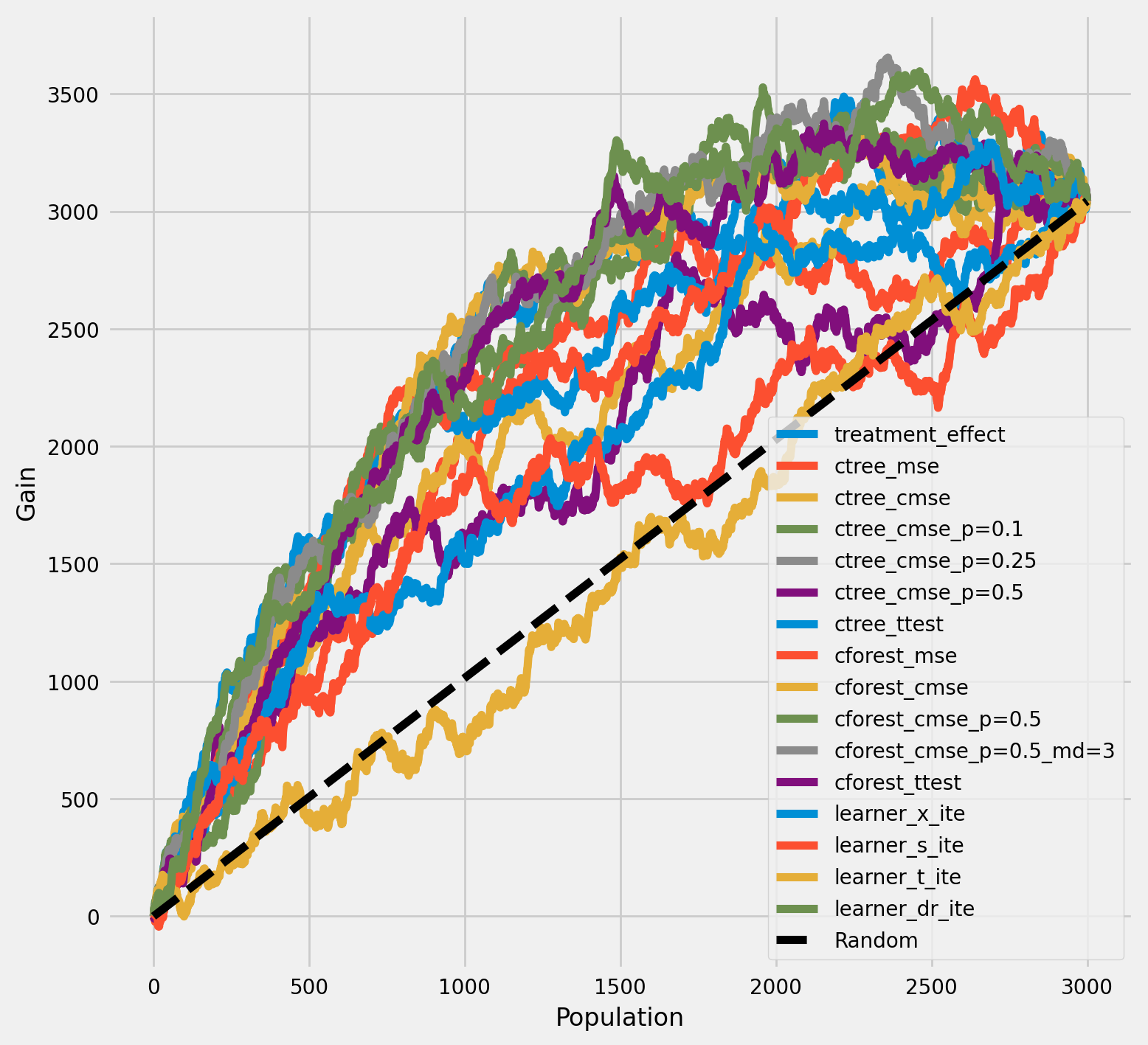
[31]:
rows = 2
cols = 7
row_idxs = np.arange(rows)
col_idxs = np.arange(cols)
ax_idxs = np.dstack(np.meshgrid(col_idxs, row_idxs)).reshape(-1, 2)
[32]:
fig, ax = plt.subplots(rows, cols, figsize=(20, 10))
plt.rcParams.update({'font.size': 10})
for ax_idx, cate, model_name in zip(ax_idxs, model_cate, model_names):
col_id, row_id = ax_idx
cur_ax = ax[row_id, col_id]
cur_ax.scatter(tau, cate, alpha=0.3)
cur_ax.plot(tau, tau, color='C2', linewidth=2)
cur_ax.set_xlabel('True ITE')
cur_ax.set_ylabel('Estimated ITE')
cur_ax.set_title(model_name)
cur_ax.set_xlim((-4, 6))
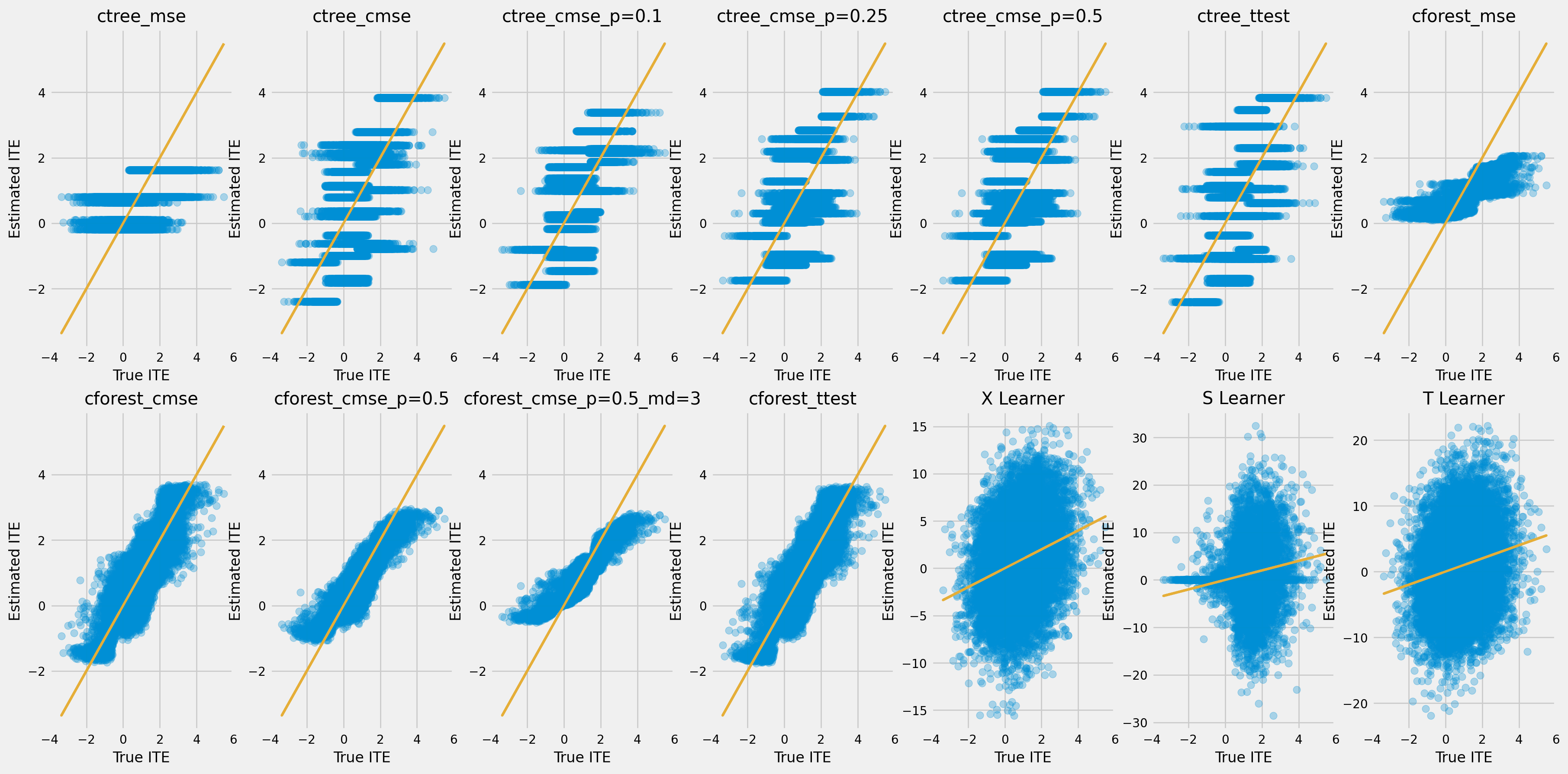
每个群体中治疗组和对照组平均结果之间的累积差异
[33]:
plot_gain(df_result,
outcome_col='outcome',
treatment_col='is_treated',
n = n_test,
figsize=(9, 9),
)

七牛图表
[34]:
plot_qini(df_result,
outcome_col='outcome',
treatment_col='is_treated',
treatment_effect_col='treatment_effect',
)
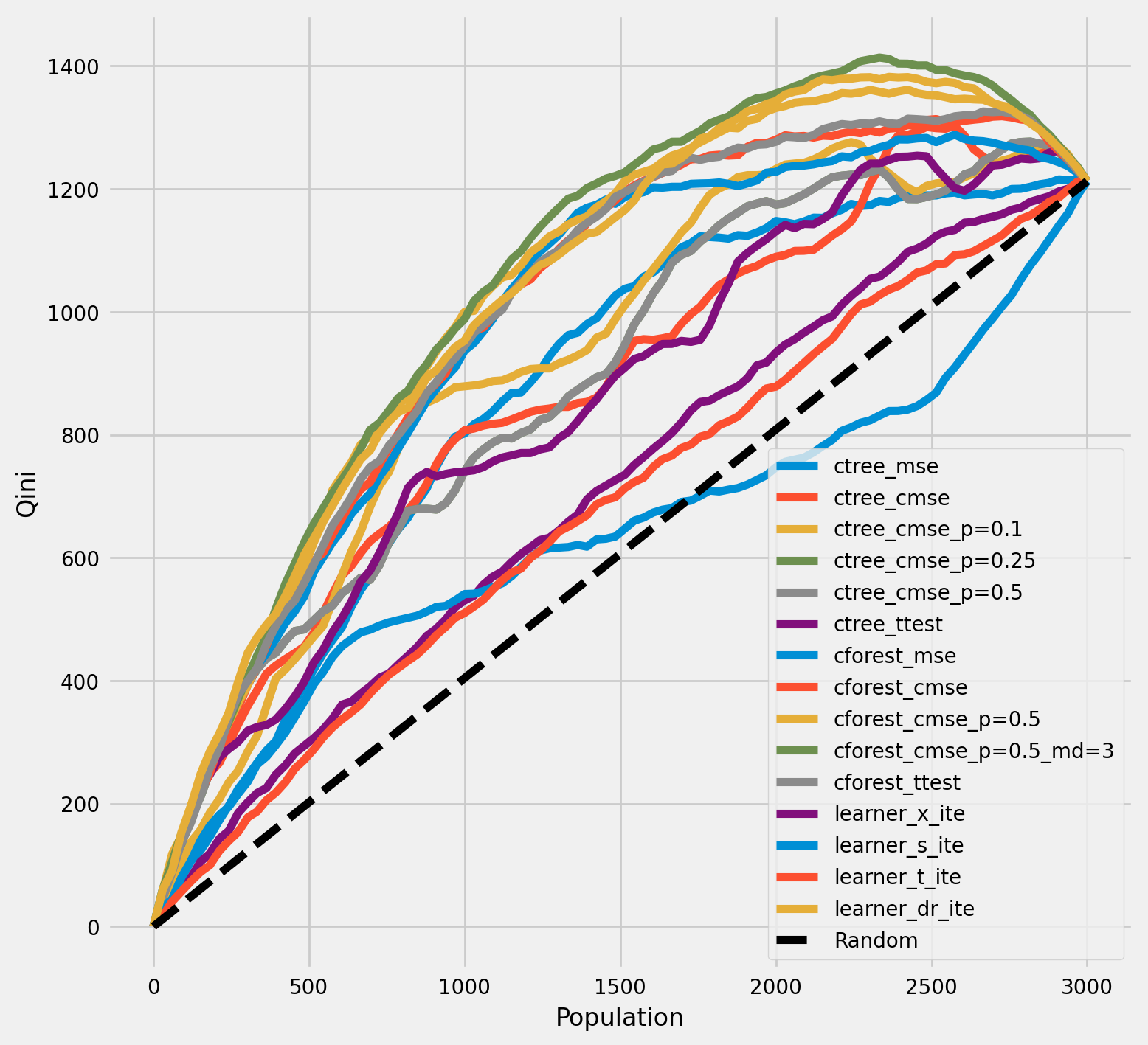
[35]:
df_qini = qini_score(df_result,
outcome_col='outcome',
treatment_col='is_treated',
treatment_effect_col='treatment_effect')
df_qini.sort_values(ascending=False)
[35]:
cforest_cmse_p=0.5_md=3 0.368213
cforest_cmse_p=0.5 0.351258
learner_dr_ite 0.349501
cforest_ttest 0.326421
cforest_cmse 0.324050
cforest_mse 0.308017
ctree_cmse_p=0.1 0.256107
ctree_cmse_p=0.25 0.228042
ctree_cmse_p=0.5 0.228042
ctree_cmse 0.218939
ctree_mse 0.214093
ctree_ttest 0.201714
learner_x_ite 0.081988
learner_t_ite 0.060148
learner_s_ite 0.023421
dtype: float64
返回结果以及估计的治疗效果
[36]:
ctree_outcomes = ctrees["ctree_mse"]["model"].predict(X_test, with_outcomes=True)
df_ctree_outcomes = pd.DataFrame(ctree_outcomes, columns=["Y0", "Y1", "ITE"])
df_ctree_outcomes.head()
[36]:
| Y0 | Y1 | ITE | |
|---|---|---|---|
| 0 | 0.770480 | 2.394923 | 1.624443 |
| 1 | 1.773314 | 2.582551 | 0.809237 |
| 2 | 0.770480 | 2.394923 | 1.624443 |
| 3 | 1.773314 | 2.582551 | 0.809237 |
| 4 | 1.773314 | 2.582551 | 0.809237 |
[37]:
cforest_outcomes = cforests["cforest_mse"]["model"].predict(X_test, with_outcomes=True)
df_cforest_outcomes = pd.DataFrame(cforest_outcomes, columns=["Y0", "Y1", "ITE"])
df_cforest_outcomes.head()
[37]:
| Y0 | Y1 | ITE | |
|---|---|---|---|
| 0 | 0.602470 | 1.519809 | 0.917338 |
| 1 | 1.839279 | 2.502261 | 0.662981 |
| 2 | 1.168312 | 2.557624 | 1.389312 |
| 3 | 1.691416 | 2.399339 | 0.707923 |
| 4 | 1.821133 | 2.495650 | 0.674517 |
个体治疗效果的Bootstrap置信区间
[38]:
alpha=0.05
tree = CausalTreeRegressor(criterion='causal_mse', control_name=0, min_samples_leaf=200, alpha=alpha)
[39]:
# For time measurements
for n_jobs in (4, mp.cpu_count() - 1):
for n_bootstraps in (10, 50, 100):
print(f"n_jobs: {n_jobs} n_bootstraps: {n_bootstraps}" )
tree.bootstrap_pool(
X=X,
treatment=w,
y=y,
n_bootstraps=n_bootstraps,
bootstrap_size=10000,
n_jobs=n_jobs,
verbose=False
)
n_jobs: 4 n_bootstraps: 10
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 10/10 [00:00<00:00, 25.17it/s]
Function: bootstrap_pool Kwargs: {'n_bootstraps': 10, 'bootstrap_size': 10000, 'n_jobs': 4, 'verbose': False} Elapsed time: 0.5051
n_jobs: 4 n_bootstraps: 50
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 50/50 [00:01<00:00, 29.83it/s]
Function: bootstrap_pool Kwargs: {'n_bootstraps': 50, 'bootstrap_size': 10000, 'n_jobs': 4, 'verbose': False} Elapsed time: 1.7102
n_jobs: 4 n_bootstraps: 100
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [00:03<00:00, 30.11it/s]
Function: bootstrap_pool Kwargs: {'n_bootstraps': 100, 'bootstrap_size': 10000, 'n_jobs': 4, 'verbose': False} Elapsed time: 3.3618
n_jobs: 7 n_bootstraps: 10
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 10/10 [00:00<00:00, 33.62it/s]
Function: bootstrap_pool Kwargs: {'n_bootstraps': 10, 'bootstrap_size': 10000, 'n_jobs': 7, 'verbose': False} Elapsed time: 0.3781
n_jobs: 7 n_bootstraps: 50
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 50/50 [00:01<00:00, 43.35it/s]
Function: bootstrap_pool Kwargs: {'n_bootstraps': 50, 'bootstrap_size': 10000, 'n_jobs': 7, 'verbose': False} Elapsed time: 1.2075
n_jobs: 7 n_bootstraps: 100
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [00:02<00:00, 44.97it/s]
Function: bootstrap_pool Kwargs: {'n_bootstraps': 100, 'bootstrap_size': 10000, 'n_jobs': 7, 'verbose': False} Elapsed time: 2.2840
[40]:
te, te_lower, te_upper = tree.fit_predict(
X=df_train[feature_names].values,
treatment=df_train["treatment"].values,
y=df_train["outcome"].values,
return_ci=True,
n_bootstraps=500,
bootstrap_size=5000,
n_jobs=mp.cpu_count() - 1,
verbose=False)
100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 500/500 [00:04<00:00, 118.14it/s]
Function: bootstrap_pool Kwargs: {'n_bootstraps': 500, 'bootstrap_size': 5000, 'n_jobs': 7, 'verbose': False} Elapsed time: 4.2891
[41]:
plt.hist(te_lower, color='red', alpha=0.3, label='lower_bound')
plt.axvline(x = 0, color = 'black', linestyle='--', lw=1, label='')
plt.legend()
plt.show()
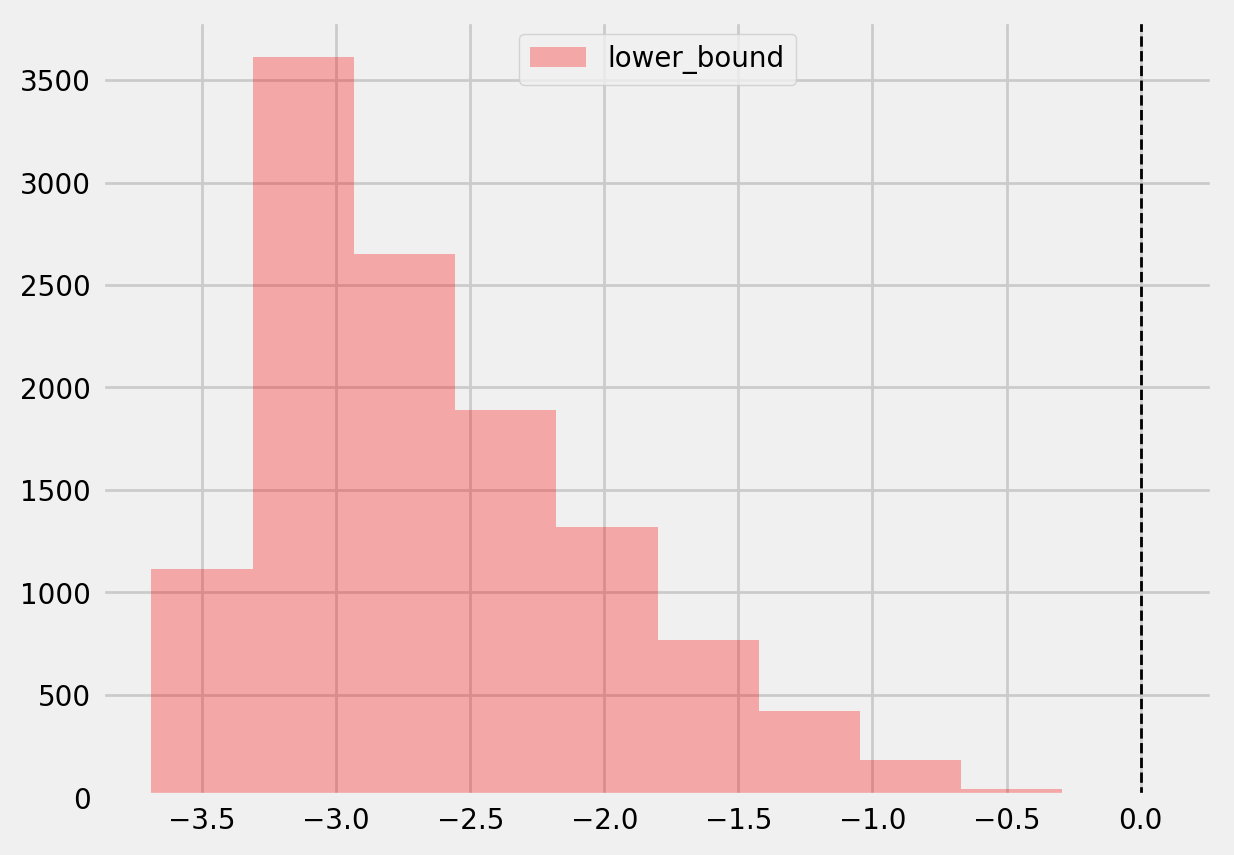
[42]:
# Significant estimates for negative and positive individual effects
# Default alpha = 0.05
bootstrap_neg = te[(te_lower < 0) & (te_upper < 0)]
bootstrap_pos = te[(te_lower > 0) & (te_upper > 0)]
print(bootstrap_neg.shape, bootstrap_pos.shape)
(0,) (3,)
[43]:
plt.hist(bootstrap_neg)
plt.title(f'Bootstrap-based subsample of significant negative ITE. alpha={alpha}')
plt.show()
plt.hist(bootstrap_pos)
plt.title(f'Bootstrap-based subsample of significant positive ITE alpha={alpha}')
plt.show()


平均处理效果
[44]:
tree = CausalTreeRegressor(criterion='causal_mse', control_name=0, min_samples_leaf=200, alpha=alpha)
te, te_lb, te_ub = tree.estimate_ate(X=X, treatment=w, y=y)
print('ATE:', te, 'Bounds:', (te_lb, te_ub ), 'alpha:', alpha)
ATE: 0.8087131266584447 Bounds: (0.8084641289076604, 0.8089621244092291) alpha: 0.05
CausalRandomForestRegressor ITE 标准差
[45]:
crforest = CausalRandomForestRegressor(criterion="causal_mse", min_samples_leaf=200,
control_name=0, n_estimators=50, n_jobs=mp.cpu_count()-1)
crforest.fit(X=df_train[feature_names].values,
treatment=df_train['treatment'].values,
y=df_train['outcome'].values
)
[45]:
CausalRandomForestRegressor(min_samples_leaf=200, n_estimators=50, n_jobs=7)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
CausalRandomForestRegressor(min_samples_leaf=200, n_estimators=50, n_jobs=7)
[46]:
crforest_te_pred = crforest.predict(df_test[feature_names])
crforest_test_var = crforest.calculate_error(X_train=df_train[feature_names].values,
X_test=df_test[feature_names].values)
crforest_test_std = np.sqrt(crforest_test_var)
[47]:
plt.hist(crforest_test_std)
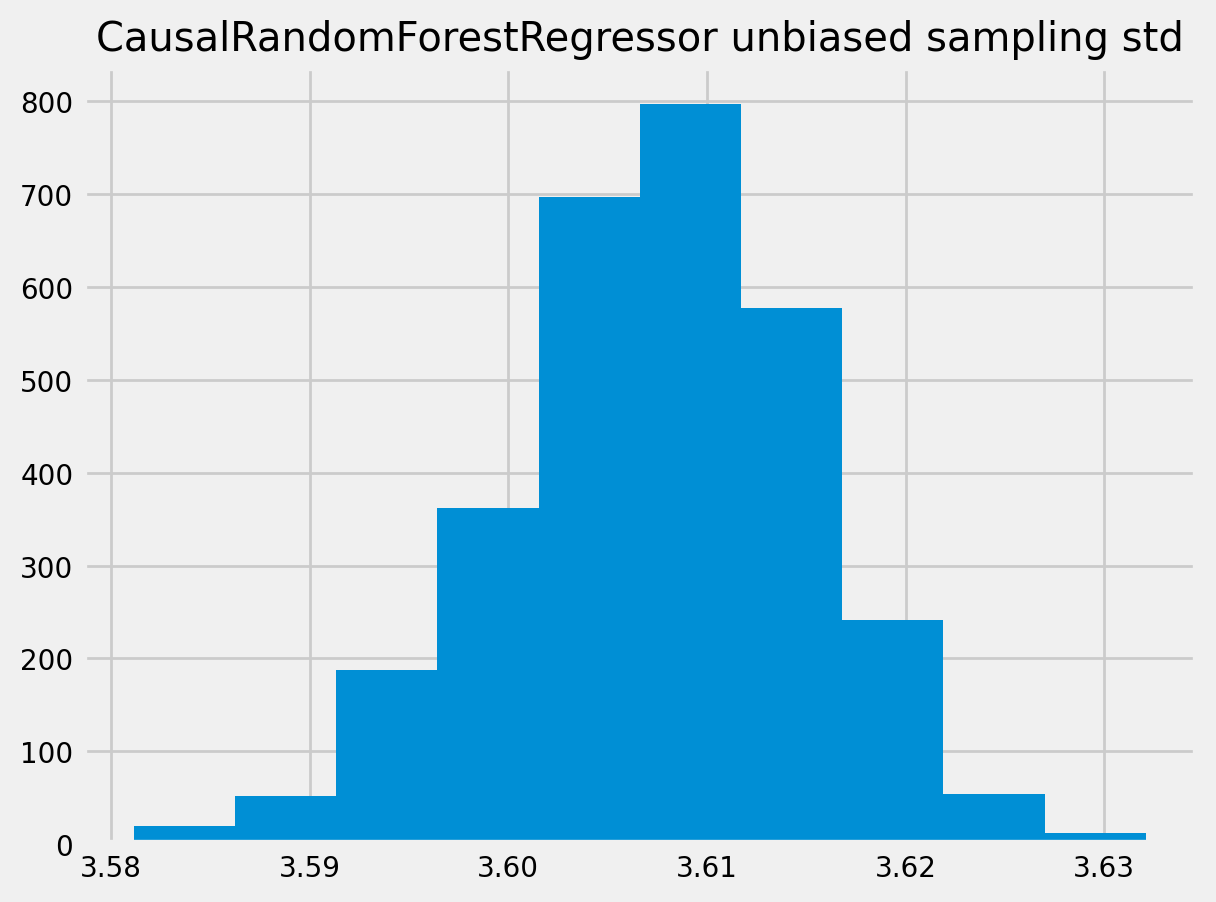
plt.title("CausalRandomForestRegressor unbiased sampling std")
plt.show()