离散变量的自动边缘化#
PyMC非常适合用于对具有离散潜在变量的模型进行采样。但如果你坚持只使用NUTS采样器,你需要以某种方式消除离散变量。最好的方法是边缘化它们,这样你可以受益于Rao-Blackwell定理,并获得参数的较低方差估计。
形式上,这个论点是这样的,采样器可以理解为近似期望值 \(\mathbb{E}_{p(x, z)}[f(x, z)]\) 对于某个函数 \(f\) 相对于分布 \(p(x, z)\)。根据全期望定律,我们知道
令 \(g(z) = \mathbb{E}_{p(x \mid z)}\left[f(x, z)\right]\),根据全变差定律,我们知道
因为期望值是针对方差计算的,所以它必须始终为正,因此我们知道
直观地说,在模型中边缘化变量可以让您使用 \(g\) 而不是 \(f\)。这种较低的方差最直接体现在较低的蒙特卡罗标准误差(mcse),间接体现在通常较高的有效样本量(ESS)。
不幸的是,进行这种计算通常是繁琐且不直观的。幸运的是,pymc-experimental 现在支持自动完成这项工作!
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pymc as pm
import pytensor.tensor as pt
import pymc_experimental as pmx
%config InlineBackend.figure_format = 'retina' # high resolution figures
az.style.use("arviz-darkgrid")
rng = np.random.default_rng(32)
作为一个激励性的例子,考虑一个高斯混合模型
高斯混合模型#
有两种方法可以指定相同的模型。一种是明确选择混合的方式。
mu = pt.as_tensor([-2.0, 2.0])
with pmx.MarginalModel() as explicit_mixture:
idx = pm.Bernoulli("idx", 0.7)
y = pm.Normal("y", mu=mu[idx], sigma=1.0)
另一种方法是使用内置的 NormalMixture 分布。在这里,混合分配在我们的模型中不是一个显式变量。第一个模型除了使用 pmx.MarginalModel 而不是 pm.Model 进行初始化之外,并没有什么特别之处。这个不同的类将允许我们在以后边缘化变量。
with pm.Model() as prebuilt_mixture:
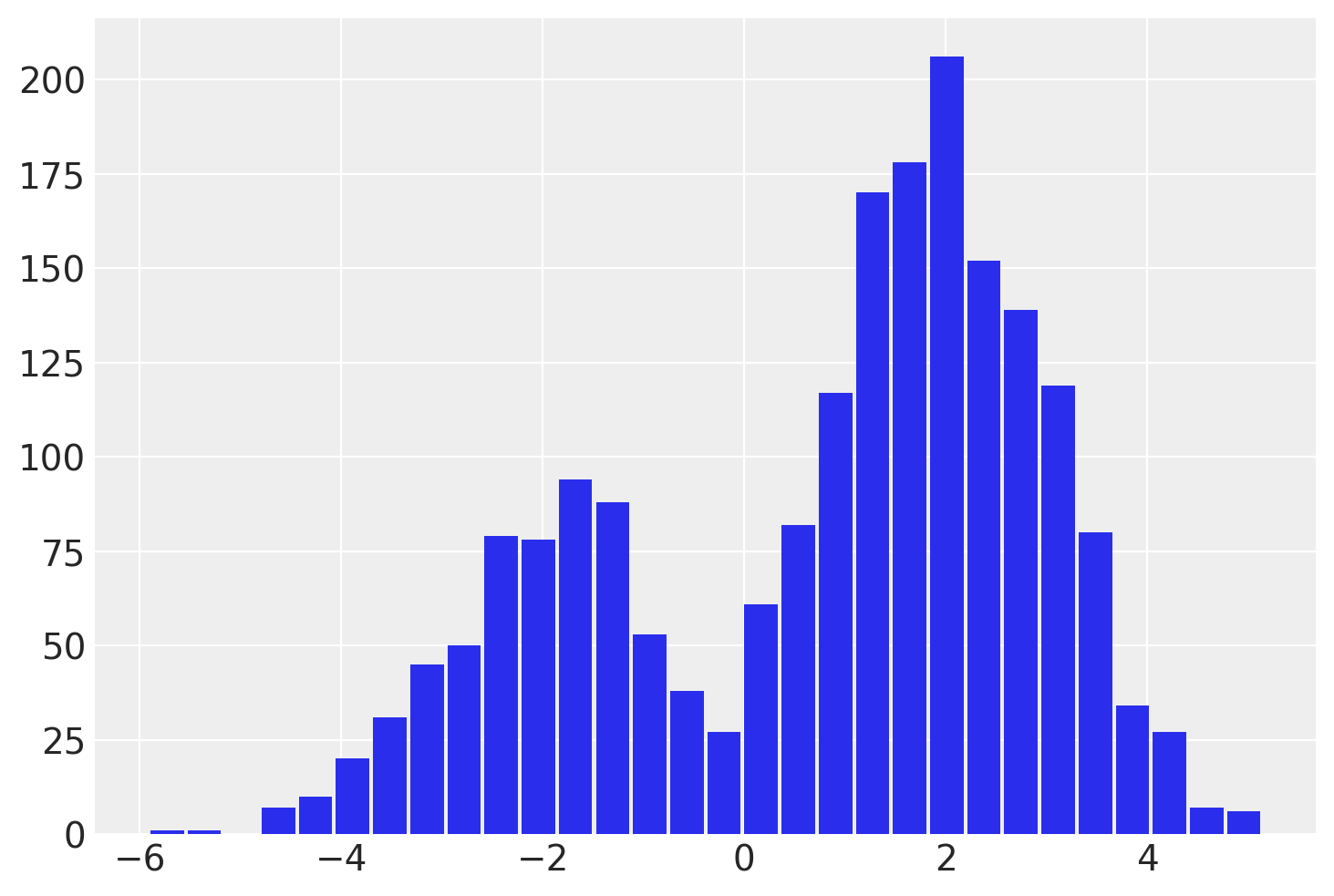
y = pm.NormalMixture("y", w=[0.3, 0.7], mu=[-2, 2])
with prebuilt_mixture:
idata = pm.sample(draws=2000, chains=4, random_seed=rng)
az.summary(idata)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 2 jobs)
NUTS: [y]
Sampling 4 chains for 1_000 tune and 2_000 draw iterations (4_000 + 8_000 draws total) took 10 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| y | 0.863 | 2.08 | -3.138 | 3.832 | 0.095 | 0.067 | 555.0 | 1829.0 | 1.01 |
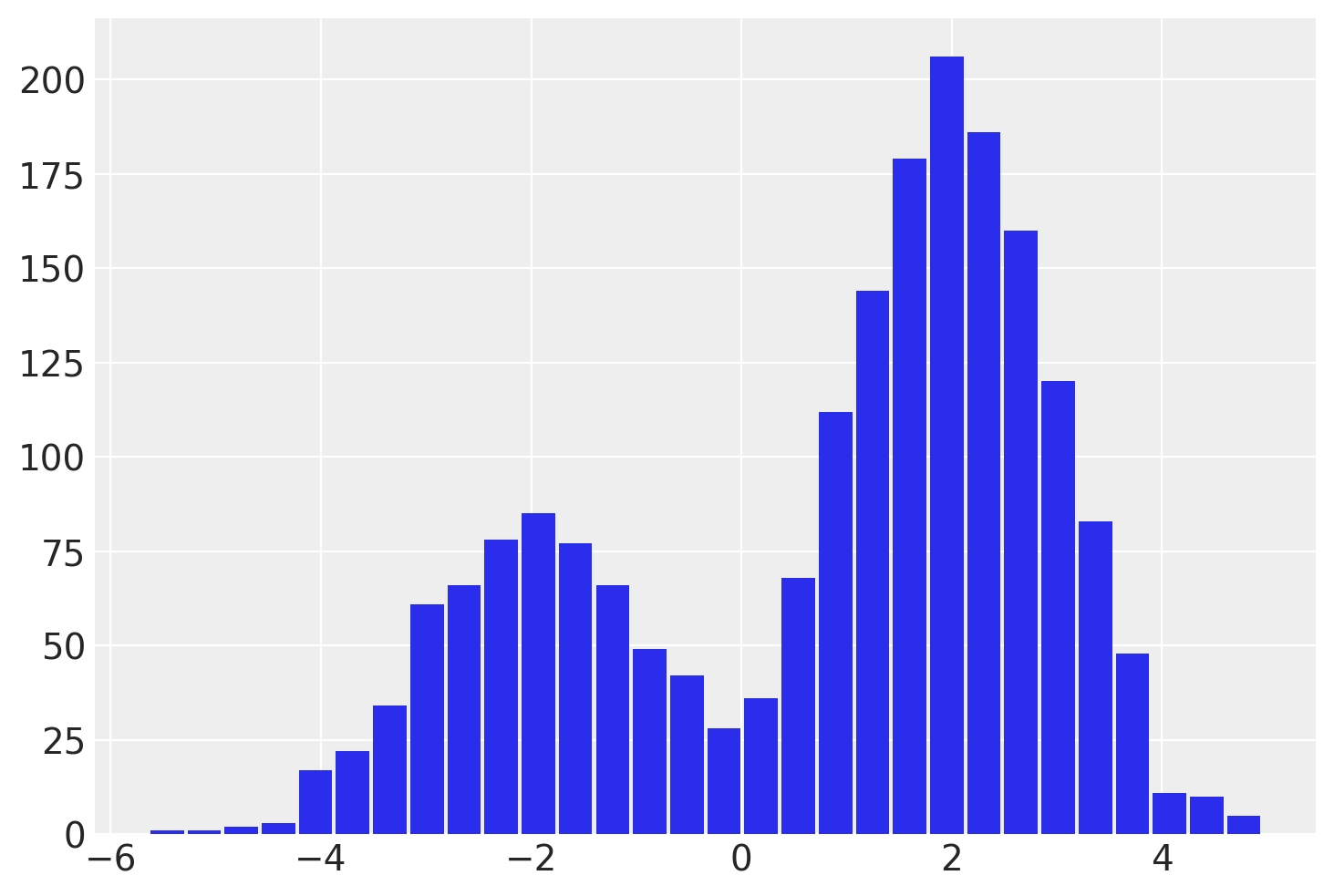
with explicit_mixture:
idata = pm.sample(draws=2000, chains=4, random_seed=rng)
az.summary(idata)
Multiprocess sampling (4 chains in 2 jobs)
CompoundStep
>BinaryGibbsMetropolis: [idx]
>NUTS: [y]
Sampling 4 chains for 1_000 tune and 2_000 draw iterations (4_000 + 8_000 draws total) took 10 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| idx | 0.718 | 0.450 | 0.000 | 1.000 | 0.028 | 0.020 | 252.0 | 252.0 | 1.02 |
| y | 0.875 | 2.068 | -3.191 | 3.766 | 0.122 | 0.087 | 379.0 | 1397.0 | 1.01 |
我们可以立即看到,边缘化模型具有更高的ESS。现在让我们边缘化选择,看看它在我们模型中有什么变化。
explicit_mixture.marginalize(["idx"])
with explicit_mixture:
idata = pm.sample(draws=2000, chains=4, random_seed=rng)
az.summary(idata)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 2 jobs)
NUTS: [y]
Sampling 4 chains for 1_000 tune and 2_000 draw iterations (4_000 + 8_000 draws total) took 10 seconds.
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| y | 0.731 | 2.102 | -3.202 | 3.811 | 0.099 | 0.07 | 567.0 | 2251.0 | 1.01 |
正如我们所见,idx 变量现在已经消失了。我们还成功使用了NUTS采样器,并且ESS有所改善。
但边际模型具有明显的优势。它仍然知道那些被边缘化的离散变量,并且我们可以获得给定其他变量的idx的后验估计。我们通过使用恢复边缘化方法来实现这一点。
explicit_mixture.recover_marginals(idata, random_seed=rng);
az.summary(idata)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| y | 0.731 | 2.102 | -3.202 | 3.811 | 0.099 | 0.070 | 567.0 | 2251.0 | 1.01 |
| idx | 0.683 | 0.465 | 0.000 | 1.000 | 0.023 | 0.016 | 420.0 | 420.0 | 1.01 |
| lp_idx[0] | -6.064 | 5.242 | -14.296 | -0.000 | 0.227 | 0.160 | 567.0 | 2251.0 | 1.01 |
| lp_idx[1] | -2.294 | 3.931 | -10.548 | -0.000 | 0.173 | 0.122 | 567.0 | 2251.0 | 1.01 |
这个 idx 变量让我们在运行NUTS采样器后恢复混合分配变量!我们可以通过从每个样本的相关 idx 中读取混合标签来拆分 y 的样本。
# fmt: off
post = idata.posterior
plt.hist(
post.where(post.idx == 0).y.values.reshape(-1),
bins=30,
rwidth=0.9,
alpha=0.75,
label='idx = 0',
)
plt.hist(
post.where(post.idx == 1).y.values.reshape(-1),
bins=30,
rwidth=0.9,
alpha=0.75,
label='idx = 1'
)
# fmt: on
plt.legend();
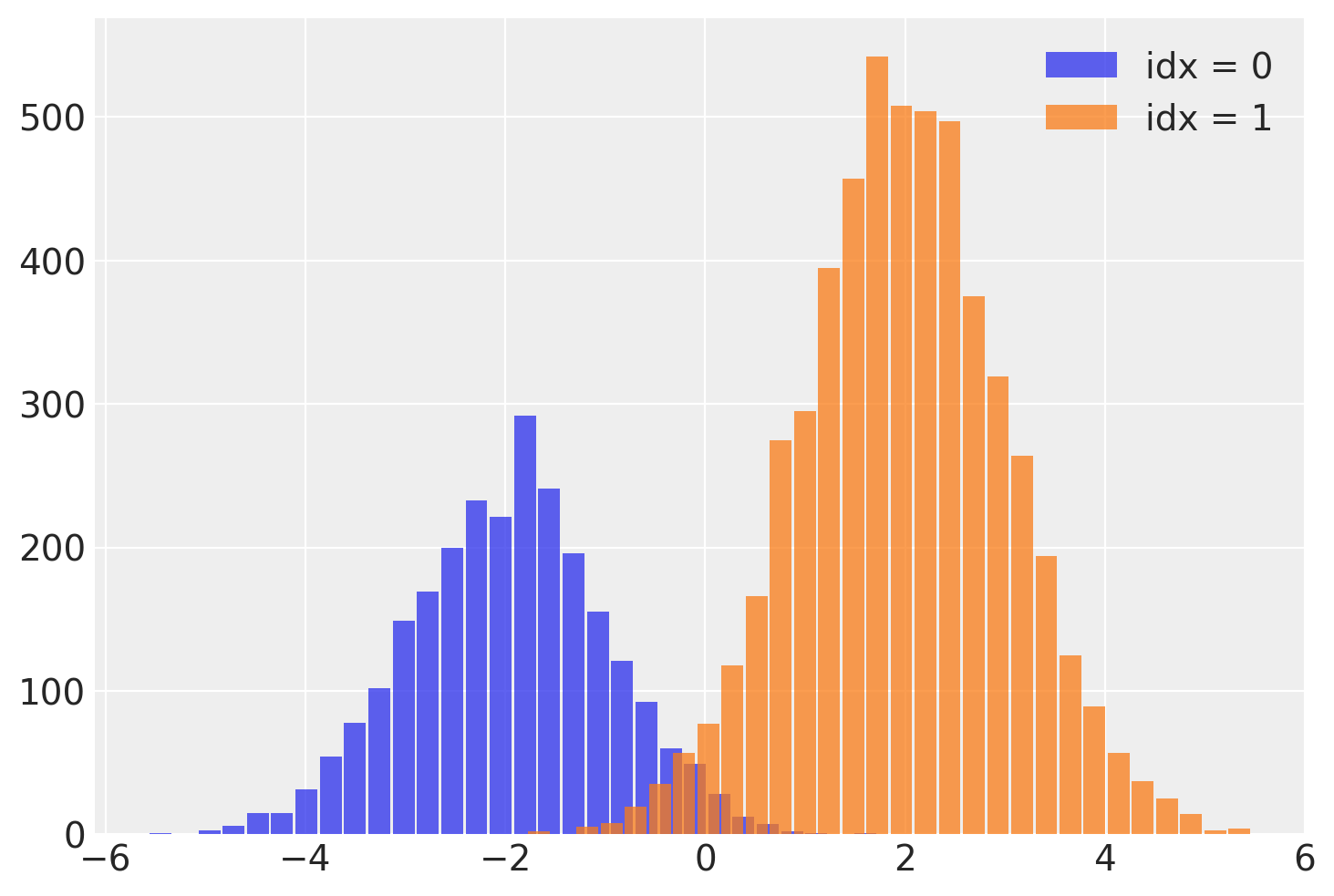
需要注意的一个重要问题是,这个离散变量的有效样本量(ESS)较低,特别是在尾部。这意味着idx可能无法很好地估计,特别是在尾部。如果这一点很重要,我建议使用lp_idx,它是给定每次迭代样本值的idx的对数概率。在下一个示例中将进一步探讨使用lp_idx的好处。
煤矿开采模型#
同样的方法也适用于煤矿开采转折点模型。煤矿开采数据集记录了1851年至1962年间英国煤矿灾难的数量。该时间序列数据集捕捉到了正在引入采矿安全法规的时期,我们尝试使用一个离散的转折点变量来估计这一事件发生的时间。
# fmt: off
disaster_data = pd.Series(
[4, 5, 4, 0, 1, 4, 3, 4, 0, 6, 3, 3, 4, 0, 2, 6,
3, 3, 5, 4, 5, 3, 1, 4, 4, 1, 5, 5, 3, 4, 2, 5,
2, 2, 3, 4, 2, 1, 3, np.nan, 2, 1, 1, 1, 1, 3, 0, 0,
1, 0, 1, 1, 0, 0, 3, 1, 0, 3, 2, 2, 0, 1, 1, 1,
0, 1, 0, 1, 0, 0, 0, 2, 1, 0, 0, 0, 1, 1, 0, 2,
3, 3, 1, np.nan, 2, 1, 1, 1, 1, 2, 4, 2, 0, 0, 1, 4,
0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1]
)
# fmt: on
years = np.arange(1851, 1962)
with pmx.MarginalModel() as disaster_model:
switchpoint = pm.DiscreteUniform("switchpoint", lower=years.min(), upper=years.max())
early_rate = pm.Exponential("early_rate", 1.0, initval=3)
late_rate = pm.Exponential("late_rate", 1.0, initval=1)
rate = pm.math.switch(switchpoint >= years, early_rate, late_rate)
disasters = pm.Poisson("disasters", rate, observed=disaster_data)
/home/zv/upstream/pymc/pymc/model/core.py:1307: RuntimeWarning: invalid value encountered in cast
data = convert_observed_data(data).astype(rv_var.dtype)
/home/zv/upstream/pymc/pymc/model/core.py:1321: ImputationWarning: Data in disasters contains missing values and will be automatically imputed from the sampling distribution.
warnings.warn(impute_message, ImputationWarning)
我们将在边缘化switchpoint变量之前和之后对模型进行采样
Multiprocess sampling (2 chains in 2 jobs)
CompoundStep
>CompoundStep
>>Metropolis: [switchpoint]
>>Metropolis: [disasters_unobserved]
>NUTS: [early_rate, late_rate]
Sampling 2 chains for 1_000 tune and 1_000 draw iterations (2_000 + 2_000 draws total) took 8 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
/home/zv/upstream/pymc-experimental/pymc_experimental/model/marginal_model.py:169: UserWarning: There are multiple dependent variables in a FiniteDiscreteMarginalRV. Their joint logp terms will be assigned to the first RV: disasters_unobserved
warnings.warn(
Multiprocess sampling (2 chains in 2 jobs)
CompoundStep
>NUTS: [early_rate, late_rate]
>Metropolis: [disasters_unobserved]
Sampling 2 chains for 1_000 tune and 1_000 draw iterations (2_000 + 2_000 draws total) took 191 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
az.summary(before_marg, var_names=["~disasters"], filter_vars="like")
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| switchpoint | 1890.224 | 2.657 | 1886.000 | 1896.000 | 0.192 | 0.136 | 201.0 | 171.0 | 1.0 |
| early_rate | 3.085 | 0.279 | 2.598 | 3.636 | 0.007 | 0.005 | 1493.0 | 1255.0 | 1.0 |
| late_rate | 0.927 | 0.114 | 0.715 | 1.143 | 0.003 | 0.002 | 1136.0 | 1317.0 | 1.0 |
az.summary(after_marg, var_names=["~disasters"], filter_vars="like")
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| early_rate | 3.077 | 0.289 | 2.529 | 3.606 | 0.007 | 0.005 | 1734.0 | 1150.0 | 1.0 |
| late_rate | 0.932 | 0.113 | 0.725 | 1.150 | 0.003 | 0.002 | 1871.0 | 1403.0 | 1.0 |
如前所述,ESS得到了极大的改善
最后,让我们恢复switchpoint变量
disaster_model.recover_marginals(after_marg);
az.summary(after_marg, var_names=["~disasters", "~lp"], filter_vars="like")
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| early_rate | 3.077 | 0.289 | 2.529 | 3.606 | 0.007 | 0.005 | 1734.0 | 1150.0 | 1.00 |
| late_rate | 0.932 | 0.113 | 0.725 | 1.150 | 0.003 | 0.002 | 1871.0 | 1403.0 | 1.00 |
| switchpoint | 1889.764 | 2.458 | 1886.000 | 1894.000 | 0.070 | 0.050 | 1190.0 | 1883.0 | 1.01 |
虽然 recover_marginals 能够对被边缘化的离散变量进行采样。每次抽样所关联的概率通常能提供一个更清晰的离散变量估计。特别是对于较低概率值的情况。通过比较采样值的直方图与对数概率图,可以最好地说明这一点。
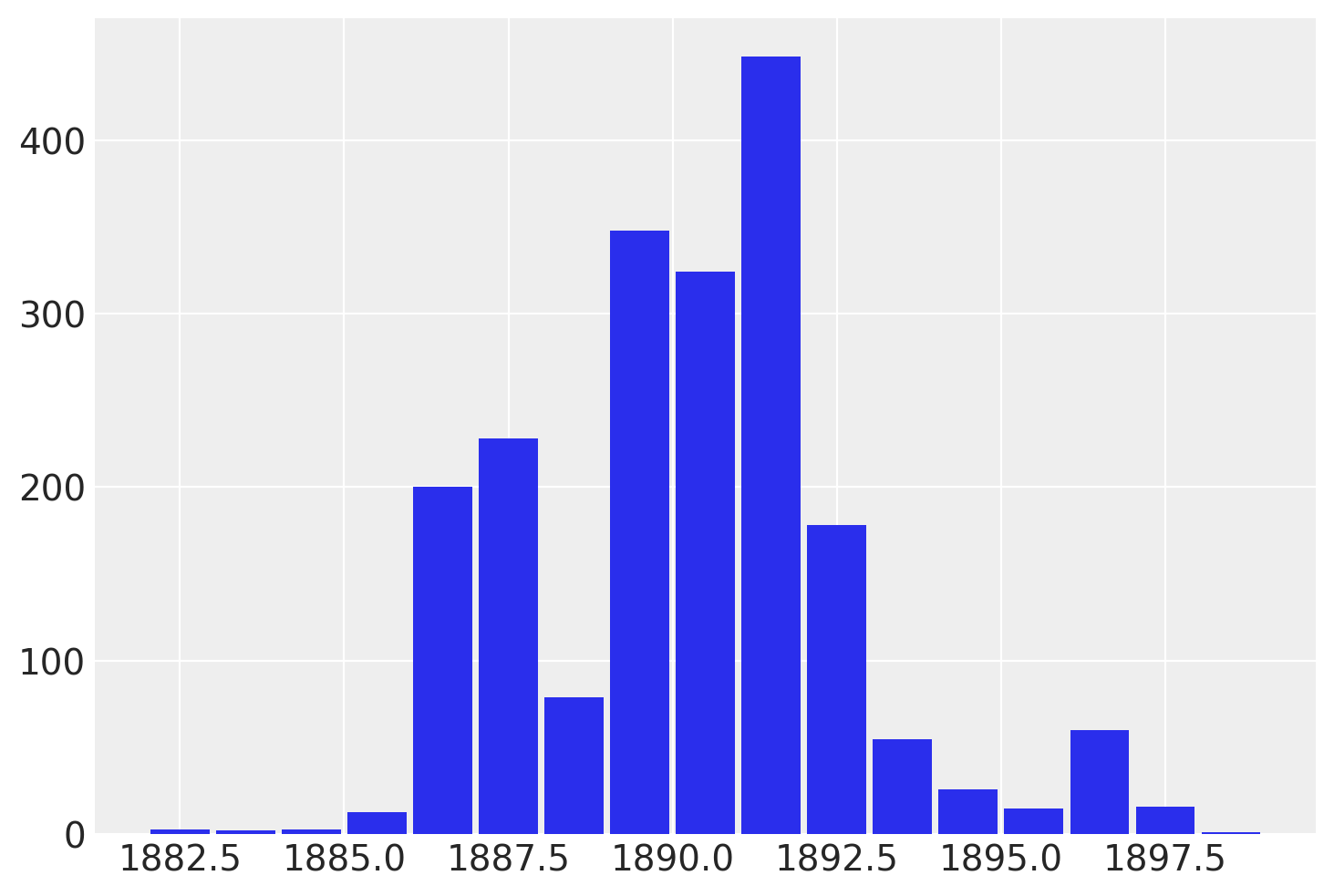
lp_switchpoint = after_marg.posterior.lp_switchpoint.mean(dim=["chain", "draw"])
x_max = years[lp_switchpoint.argmax()]
plt.scatter(years, lp_switchpoint)
plt.axvline(x=x_max, c="orange")
plt.xlabel(r"$\mathrm{year}$")
plt.ylabel(r"$\log p(\mathrm{switchpoint}=\mathrm{year})$");
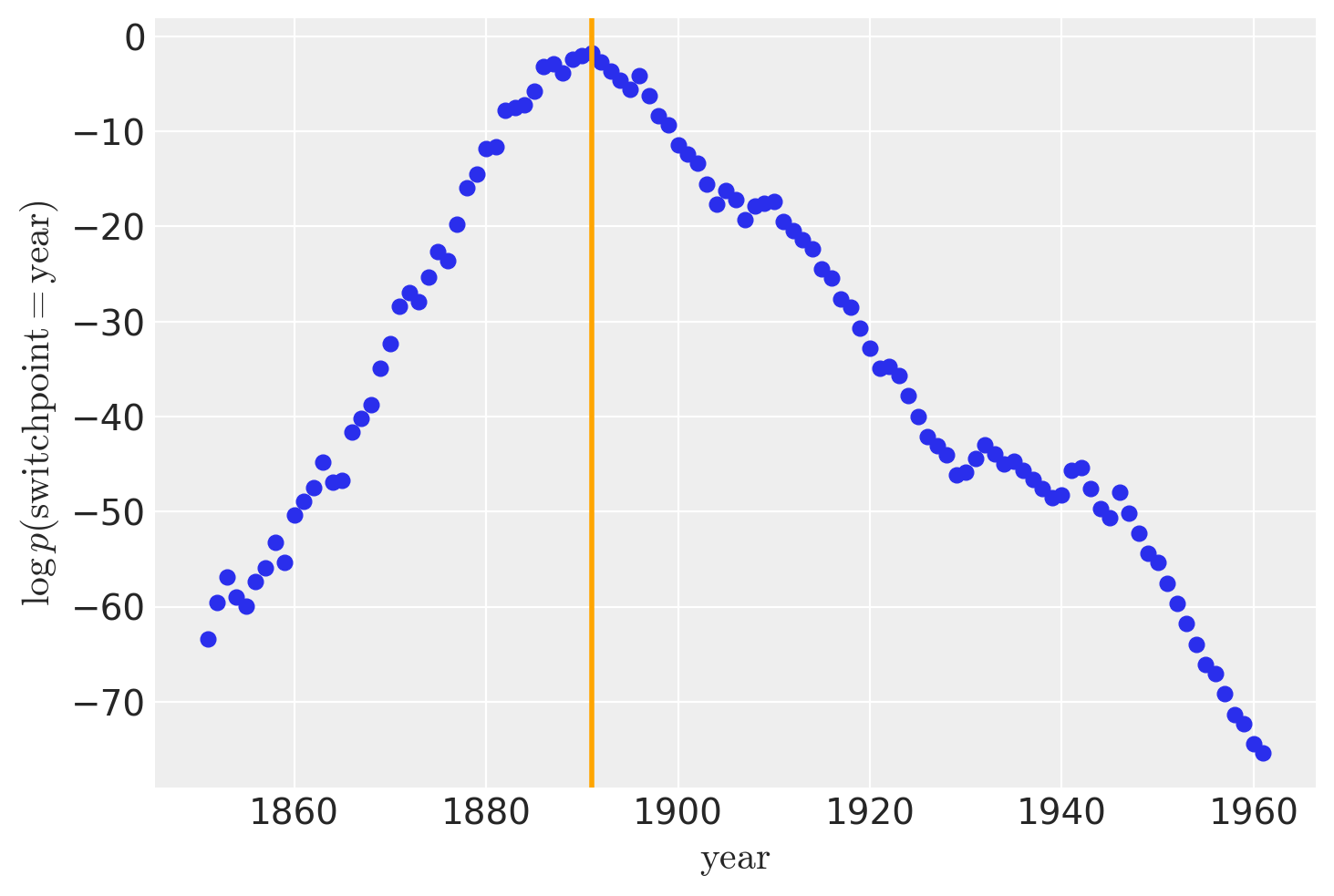
通过绘制采样值的直方图而不是直接处理对数概率,我们对底层离散分布的探索变得更加嘈杂和不完整。
参考资料#
水印#
%load_ext watermark
%watermark -n -u -v -iv -w -p pytensor,xarray
Last updated: Sat Feb 10 2024
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.20.0
pytensor: 2.18.6
xarray : 2023.11.0
pymc : 5.11
numpy : 1.26.3
pytensor : 2.18.6
pymc_experimental: 0.0.15
arviz : 0.17.0
pandas : 2.1.4
matplotlib : 3.8.2
Watermark: 2.4.3