重新参数化Weibull加速失效时间模型#
import arviz as az
import numpy as np
import pymc as pm
import pytensor.tensor as pt
import statsmodels.api as sm
print(f"Running on PyMC v{pm.__version__}")
Running on PyMC v5.0.1+42.g99dd7158
%config InlineBackend.figure_format = 'retina'
RANDOM_SEED = 8927
np.random.seed(RANDOM_SEED)
az.style.use("arviz-darkgrid")
数据集#
之前的关于贝叶斯参数生存分析的示例笔记本介绍了两种不同的加速失效时间(AFT)模型:Weibull和线性对数模型。在本笔记本中,我们介绍了Weibull AFT模型的三种不同参数化方法。
我们将使用的数据集是 flchain R 数据集,该数据集来自一项医学研究,研究血清游离轻链 (FLC) 对寿命的影响。通过运行以下命令阅读数据的完整文档:
print(sm.datasets.get_rdataset(package='survival', dataname='flchain').__doc__)。
# Fetch and clean data
data = (
sm.datasets.get_rdataset(package="survival", dataname="flchain")
.data.sample(500) # Limit ourselves to 500 observations
.reset_index(drop=True)
)
y = data.futime.values
censored = ~data["death"].values.astype(bool)
y[:5]
array([ 975, 2272, 138, 4262, 4928])
censored[:5]
array([False, True, False, True, True])
使用 pm.Potential#
我们在对截尾数据进行建模时遇到了一个独特的问题。严格来说,我们没有任何关于截尾值的数据:我们只知道被截尾的数量。我们如何将这些信息纳入我们的模型中?
一种方法是利用pm.Potential。PyMC2 文档很好地解释了它的用法。本质上,声明pm.Potential('x', logp)会将logp添加到模型的对数似然中。
参数化 1#
这种参数化是威布尔生存函数的一种直观、直接的参数化方法。这可能是人们首先想到的参数化方法。
def weibull_lccdf(x, alpha, beta):
"""Log complementary cdf of Weibull distribution."""
return -((x / beta) ** alpha)
with pm.Model() as model_1:
alpha_sd = 10.0
mu = pm.Normal("mu", mu=0, sigma=100)
alpha_raw = pm.Normal("a0", mu=0, sigma=0.1)
alpha = pm.Deterministic("alpha", pt.exp(alpha_sd * alpha_raw))
beta = pm.Deterministic("beta", pt.exp(mu / alpha))
y_obs = pm.Weibull("y_obs", alpha=alpha, beta=beta, observed=y[~censored])
y_cens = pm.Potential("y_cens", weibull_lccdf(y[censored], alpha, beta))
with model_1:
# Change init to avoid divergences
data_1 = pm.sample(target_accept=0.9, init="adapt_diag")
Auto-assigning NUTS sampler...
Initializing NUTS using adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [mu, a0]
100.00% [8000/8000 00:09<00:00 Sampling 4 chains, 0 divergences]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 10 seconds.
az.plot_trace(data_1, var_names=["alpha", "beta"])
array([[<AxesSubplot: title={'center': 'alpha'}>,
<AxesSubplot: title={'center': 'alpha'}>],
[<AxesSubplot: title={'center': 'beta'}>,
<AxesSubplot: title={'center': 'beta'}>]], dtype=object)
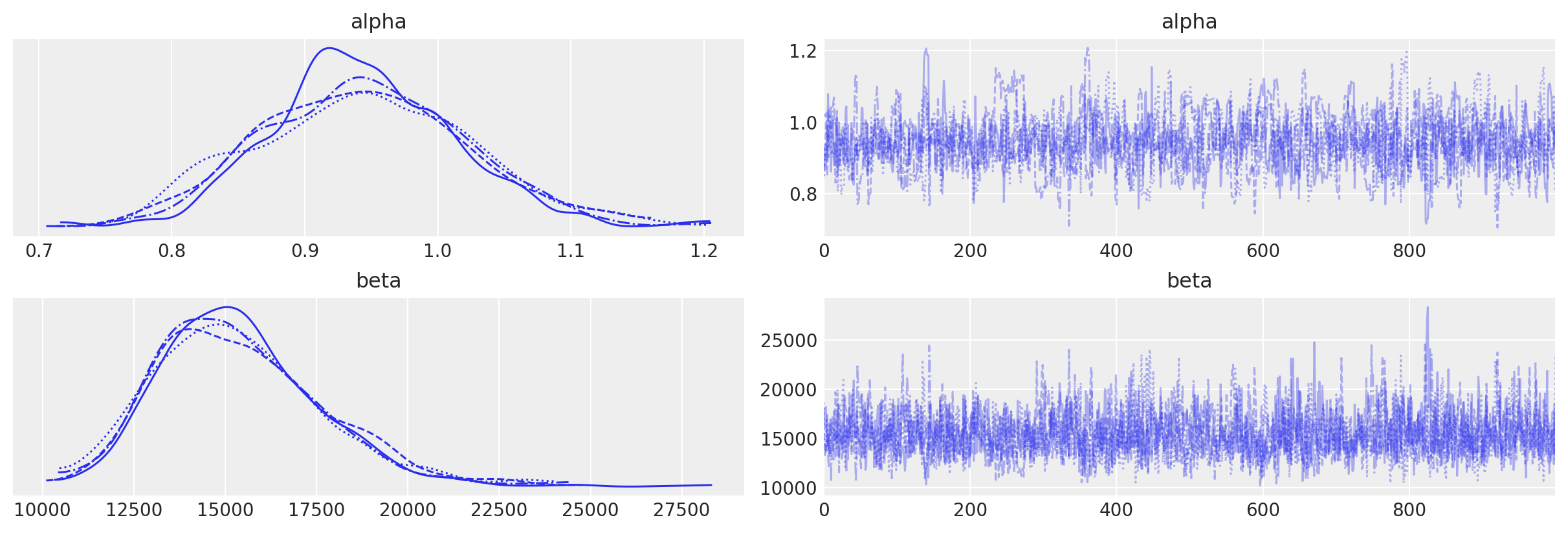
az.summary(data_1, var_names=["alpha", "beta"], round_to=2)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.94 | 0.08 | 0.80 | 1.08 | 0.00 | 0.00 | 735.35 | 701.64 | 1.01 |
| beta | 15386.70 | 2269.14 | 11442.57 | 19562.58 | 65.38 | 46.48 | 1228.87 | 1682.03 | 1.00 |
参数化 2#
需要注意的是,令人困惑的是,alpha 现在被称为 r,而 alpha 表示一个先验;我们保持这种符号以忠实于Stan中的原始实现。在这种参数化中,我们仍然对相同的参数 alpha(现在是 r)和 beta 进行建模。
更多信息,请参阅 这个 Stan 示例模型 和 相应的文档。
with pm.Model() as model_2:
alpha = pm.Normal("alpha", mu=0, sigma=10)
r = pm.Gamma("r", alpha=1, beta=0.001, testval=0.25)
beta = pm.Deterministic("beta", pt.exp(-alpha / r))
y_obs = pm.Weibull("y_obs", alpha=r, beta=beta, observed=y[~censored])
y_cens = pm.Potential("y_cens", weibull_lccdf(y[censored], r, beta))
/tmp/ipykernel_915/3602845657.py:3: FutureWarning: The `testval` argument is deprecated; use `initval`.
r = pm.Gamma("r", alpha=1, beta=0.001, testval=0.25)
with model_2:
# Increase target_accept to avoid divergences
data_2 = pm.sample(target_accept=0.9)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, r]
100.00% [8000/8000 00:10<00:00 Sampling 4 chains, 0 divergences]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 10 seconds.
az.plot_trace(data_2, var_names=["r", "beta"])
array([[<AxesSubplot: title={'center': 'r'}>,
<AxesSubplot: title={'center': 'r'}>],
[<AxesSubplot: title={'center': 'beta'}>,
<AxesSubplot: title={'center': 'beta'}>]], dtype=object)
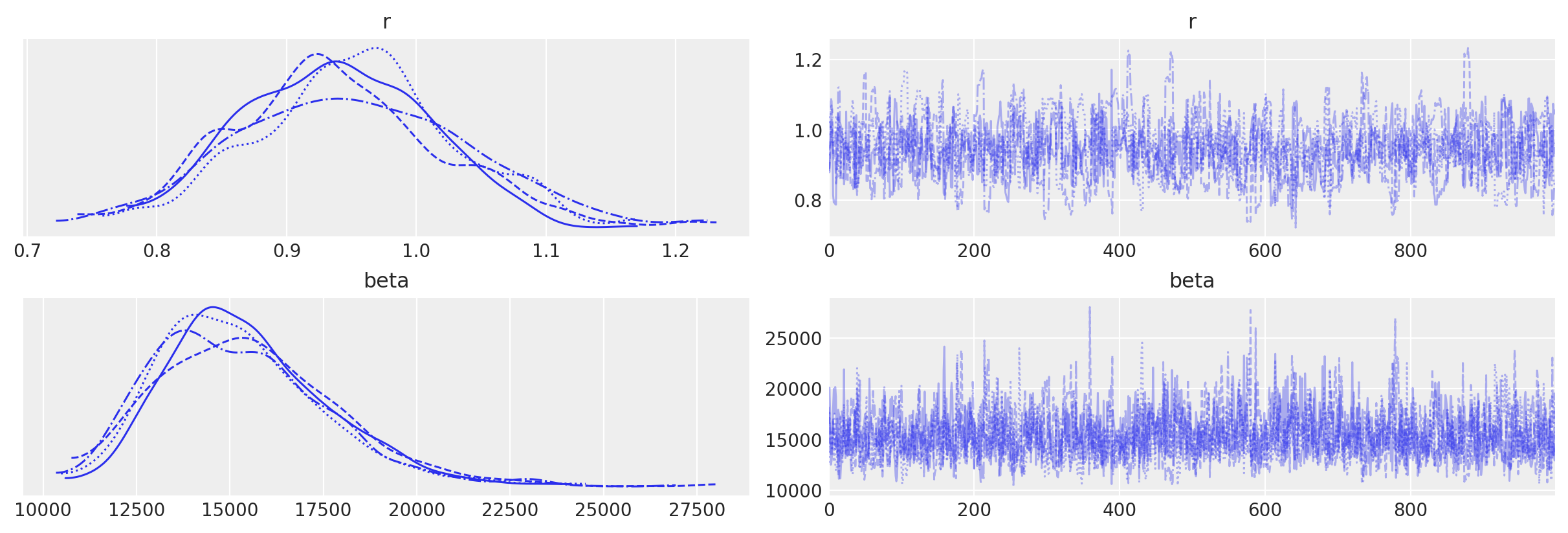
az.summary(data_2, var_names=["r", "beta"], round_to=2)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| r | 0.94 | 0.08 | 0.80 | 1.10 | 0.0 | 0.00 | 702.52 | 671.31 | 1.01 |
| beta | 15377.49 | 2313.49 | 11423.58 | 19710.63 | 65.1 | 46.47 | 1284.63 | 1696.35 | 1.00 |
参数化 3#
在这个参数化中,我们使用Gumbel分布来建模对数线性误差分布,而不是直接建模生存函数。更多信息,请参见这篇博客文章。
with model_3:
# Change init to avoid divergences
data_3 = pm.sample(init="adapt_diag")
Auto-assigning NUTS sampler...
Initializing NUTS using adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [s, gamma]
100.00% [8000/8000 00:03<00:00 Sampling 4 chains, 0 divergences]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 4 seconds.
az.plot_trace(data_3)
array([[<AxesSubplot: title={'center': 'gamma'}>,
<AxesSubplot: title={'center': 'gamma'}>],
[<AxesSubplot: title={'center': 's'}>,
<AxesSubplot: title={'center': 's'}>]], dtype=object)
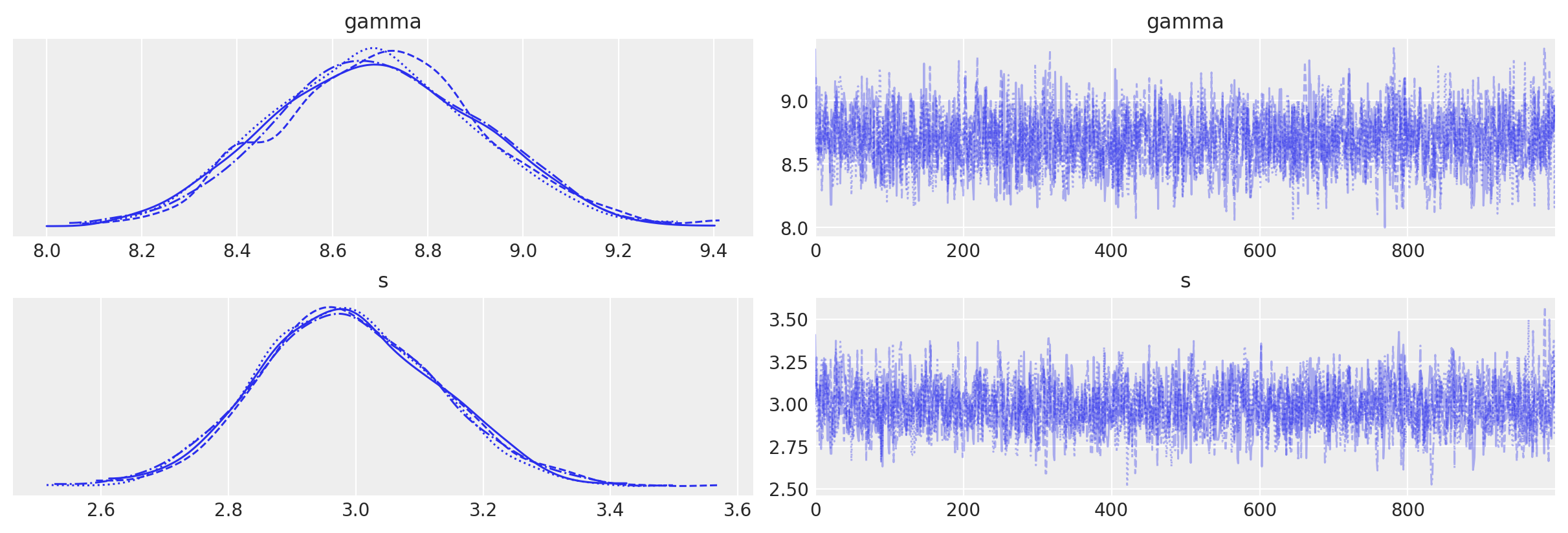
az.summary(data_3, round_to=2)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| gamma | 8.69 | 0.22 | 8.31 | 9.11 | 0.0 | 0.0 | 2233.04 | 2305.13 | 1.0 |
| s | 2.99 | 0.14 | 2.74 | 3.26 | 0.0 | 0.0 | 2067.28 | 2328.40 | 1.0 |