GLM: 分层回归模型上的小批量ADVI#
与高斯混合模型不同,(分层)回归模型具有自变量。这些变量影响似然函数,但不是随机变量。在使用小批量时,我们应该注意这一点。
%env PYTENSOR_FLAGS=device=cpu, floatX=float32, warn_float64=ignore
import os
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pymc as pm
import pytensor
import pytensor.tensor as pt
import seaborn as sns
from scipy import stats
print(f"Running on PyMC v{pm.__version__}")
env: PYTENSOR_FLAGS=device=cpu, floatX=float32, warn_float64=ignore
Running on PyMC v5.0.1
%config InlineBackend.figure_format = 'retina'
RANDOM_SEED = 8927
rng = np.random.default_rng(RANDOM_SEED)
az.style.use("arviz-darkgrid")
try:
data = pd.read_csv(os.path.join("..", "data", "radon.csv"))
except FileNotFoundError:
data = pd.read_csv(pm.get_data("radon.csv"))
data
| Unnamed: 0 | idnum | state | state2 | stfips | zip | region | typebldg | floor | room | ... | pcterr | adjwt | dupflag | zipflag | cntyfips | county | fips | Uppm | county_code | log_radon | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 5081.0 | MN | MN | 27.0 | 55735 | 5.0 | 1.0 | 1.0 | 3.0 | ... | 9.7 | 1146.499190 | 1.0 | 0.0 | 1.0 | AITKIN | 27001.0 | 0.502054 | 0 | 0.832909 |
| 1 | 1 | 5082.0 | MN | MN | 27.0 | 55748 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 14.5 | 471.366223 | 0.0 | 0.0 | 1.0 | AITKIN | 27001.0 | 0.502054 | 0 | 0.832909 |
| 2 | 2 | 5083.0 | MN | MN | 27.0 | 55748 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 9.6 | 433.316718 | 0.0 | 0.0 | 1.0 | AITKIN | 27001.0 | 0.502054 | 0 | 1.098612 |
| 3 | 3 | 5084.0 | MN | MN | 27.0 | 56469 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 24.3 | 461.623670 | 0.0 | 0.0 | 1.0 | AITKIN | 27001.0 | 0.502054 | 0 | 0.095310 |
| 4 | 4 | 5085.0 | MN | MN | 27.0 | 55011 | 3.0 | 1.0 | 0.0 | 4.0 | ... | 13.8 | 433.316718 | 0.0 | 0.0 | 3.0 | ANOKA | 27003.0 | 0.428565 | 1 | 1.163151 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 914 | 914 | 5995.0 | MN | MN | 27.0 | 55363 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 4.5 | 1146.499190 | 0.0 | 0.0 | 171.0 | WRIGHT | 27171.0 | 0.913909 | 83 | 1.871802 |
| 915 | 915 | 5996.0 | MN | MN | 27.0 | 55376 | 5.0 | 1.0 | 0.0 | 7.0 | ... | 8.3 | 1105.956867 | 0.0 | 0.0 | 171.0 | WRIGHT | 27171.0 | 0.913909 | 83 | 1.526056 |
| 916 | 916 | 5997.0 | MN | MN | 27.0 | 55376 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 5.2 | 1214.922779 | 0.0 | 0.0 | 171.0 | WRIGHT | 27171.0 | 0.913909 | 83 | 1.629241 |
| 917 | 917 | 5998.0 | MN | MN | 27.0 | 56297 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 9.6 | 1177.377355 | 0.0 | 0.0 | 173.0 | YELLOW MEDICINE | 27173.0 | 1.426590 | 84 | 1.335001 |
| 918 | 918 | 5999.0 | MN | MN | 27.0 | 56297 | 5.0 | 1.0 | 0.0 | 4.0 | ... | 8.0 | 1214.922779 | 0.0 | 0.0 | 173.0 | YELLOW MEDICINE | 27173.0 | 1.426590 | 84 | 1.098612 |
919行 × 30列
county_idx = data["county_code"].values
floor_idx = data["floor"].values
log_radon_idx = data["log_radon"].values
coords = {"counties": data.county.unique()}
在这里,log_radon_idx_t 是一个因变量,而 floor_idx_t 和 county_idx_t 决定了自变量。
log_radon_idx_t = pm.Minibatch(log_radon_idx, batch_size=100)
floor_idx_t = pm.Minibatch(floor_idx, batch_size=100)
county_idx_t = pm.Minibatch(county_idx, batch_size=100)
with pm.Model(coords=coords) as hierarchical_model:
# Hyperpriors for group nodes
mu_a = pm.Normal("mu_alpha", mu=0.0, sigma=100**2)
sigma_a = pm.Uniform("sigma_alpha", lower=0, upper=100)
mu_b = pm.Normal("mu_beta", mu=0.0, sigma=100**2)
sigma_b = pm.Uniform("sigma_beta", lower=0, upper=100)
每个县的截距,分布在组均值 mu_a 周围。在上面的例子中,我们只是将 mu 和 sd 设置为固定值,而在这里我们为所有的 a 和 b(它们是与示例中唯一县的数量相同长度的向量)插入了一个共同的组分布。
氡水平的模型预测 a[county_idx] 转换为 a[0, 0, 0, 1, 1, ...],因此我们将一个县的多个家庭测量值与其系数相关联。
最后,我们指定似然函数:
with hierarchical_model:
# Model error
eps = pm.Uniform("eps", lower=0, upper=100)
# Data likelihood
radon_like = pm.Normal(
"radon_like", mu=radon_est, sigma=eps, observed=log_radon_idx_t, total_size=len(data)
)
随机变量 radon_like,与 log_radon_idx_t 相关联,应传递给函数以表示在似然项中的观测值。
另一方面,minibatches 应该包括上述三个变量。
然后,使用小批量运行ADVI。
with hierarchical_model:
approx = pm.fit(100_000, callbacks=[pm.callbacks.CheckParametersConvergence(tolerance=1e-4)])
idata_advi = approx.sample(500)
/home/fonnesbeck/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/logprob/joint_logprob.py:167: UserWarning: Found a random variable that was neither among the observations nor the conditioned variables: [integers_rv{0, (0, 0), int64, False}.0, integers_rv{0, (0, 0), int64, False}.out]
warnings.warn(
/home/fonnesbeck/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/logprob/joint_logprob.py:167: UserWarning: Found a random variable that was neither among the observations nor the conditioned variables: [integers_rv{0, (0, 0), int64, False}.0, integers_rv{0, (0, 0), int64, False}.out]
warnings.warn(
/home/fonnesbeck/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/logprob/joint_logprob.py:167: UserWarning: Found a random variable that was neither among the observations nor the conditioned variables: [integers_rv{0, (0, 0), int64, False}.0, integers_rv{0, (0, 0), int64, False}.out]
warnings.warn(
/home/fonnesbeck/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/logprob/joint_logprob.py:167: UserWarning: Found a random variable that was neither among the observations nor the conditioned variables: [integers_rv{0, (0, 0), int64, False}.0, integers_rv{0, (0, 0), int64, False}.out]
warnings.warn(
11.50% [11499/100000 00:03<00:27 Average Loss = 263.87]
检查ELBO的轨迹并与MCMC的结果进行比较。
plt.plot(approx.hist);
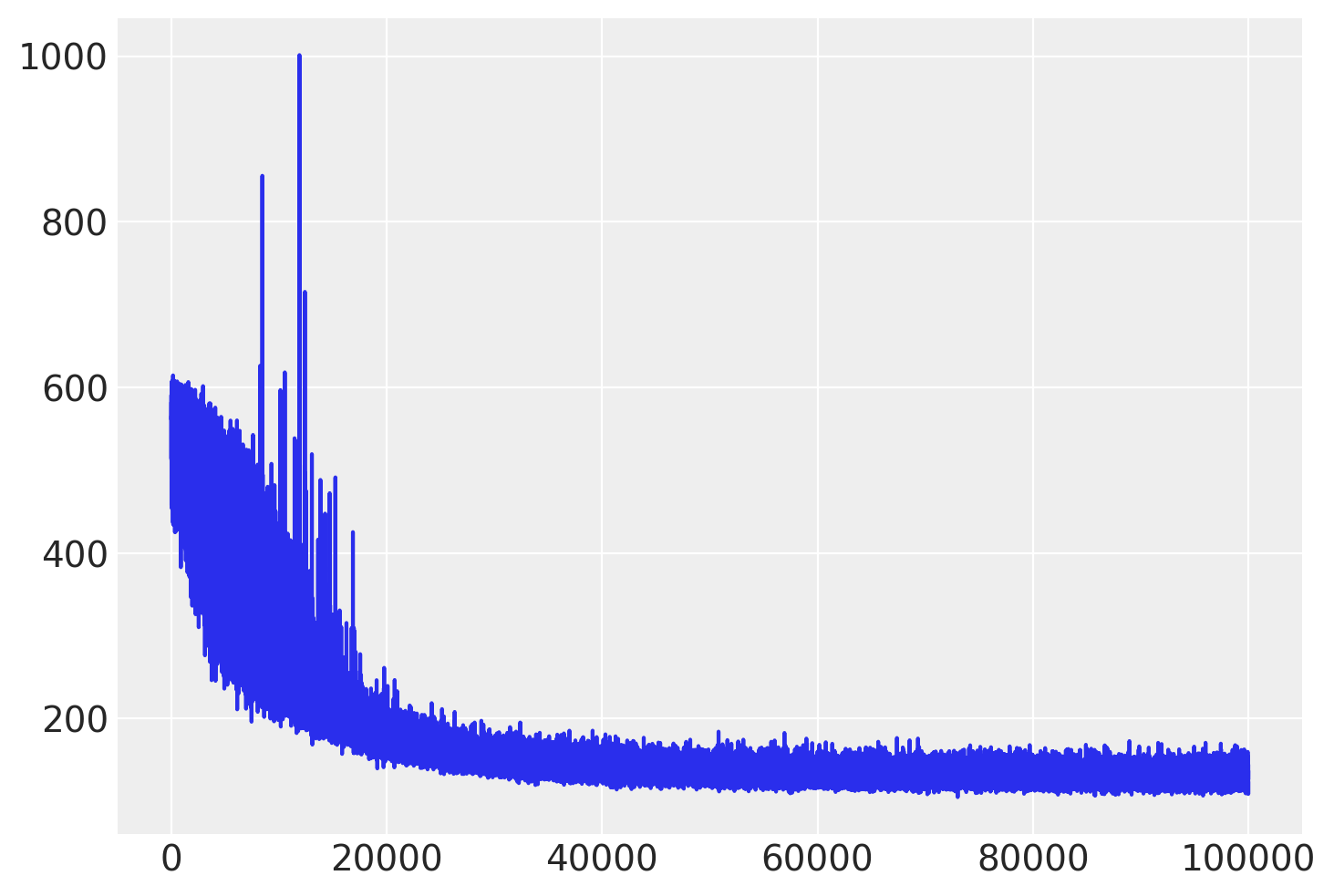
我们可以从均值场近似中提取协方差矩阵,并将其用作NUTS算法的缩放矩阵。
scaling = approx.cov.eval()
此外,我们可以生成样本(每个链一个),用作采样器的起始点。
n_chains = 4
sample = approx.sample(return_inferencedata=False, size=n_chains)
start_dict = list(sample[i] for i in range(n_chains))
# Inference button (TM)!
with pm.Model(coords=coords):
mu_a = pm.Normal("mu_alpha", mu=0.0, sigma=100**2)
sigma_a = pm.Uniform("sigma_alpha", lower=0, upper=100)
mu_b = pm.Normal("mu_beta", mu=0.0, sigma=100**2)
sigma_b = pm.Uniform("sigma_beta", lower=0, upper=100)
a = pm.Normal("alpha", mu=mu_a, sigma=sigma_a, dims="counties")
b = pm.Normal("beta", mu=mu_b, sigma=sigma_b, dims="counties")
# Model error
eps = pm.Uniform("eps", lower=0, upper=100)
radon_est = a[county_idx] + b[county_idx] * floor_idx
radon_like = pm.Normal("radon_like", mu=radon_est, sigma=eps, observed=log_radon_idx)
# essentially, this is what init='advi' does
step = pm.NUTS(scaling=scaling, is_cov=True)
hierarchical_trace = pm.sample(draws=2000, step=step, chains=n_chains, initvals=start_dict)
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [mu_alpha, sigma_alpha, mu_beta, sigma_beta, alpha, beta, eps]
100.00% [8000/8000 08:57<00:00 Sampling 4 chains, 0 divergences]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 538 seconds.
az.plot_density(
data=[idata_advi, hierarchical_trace],
var_names=["~alpha", "~beta"],
data_labels=["ADVI", "NUTS"],
);
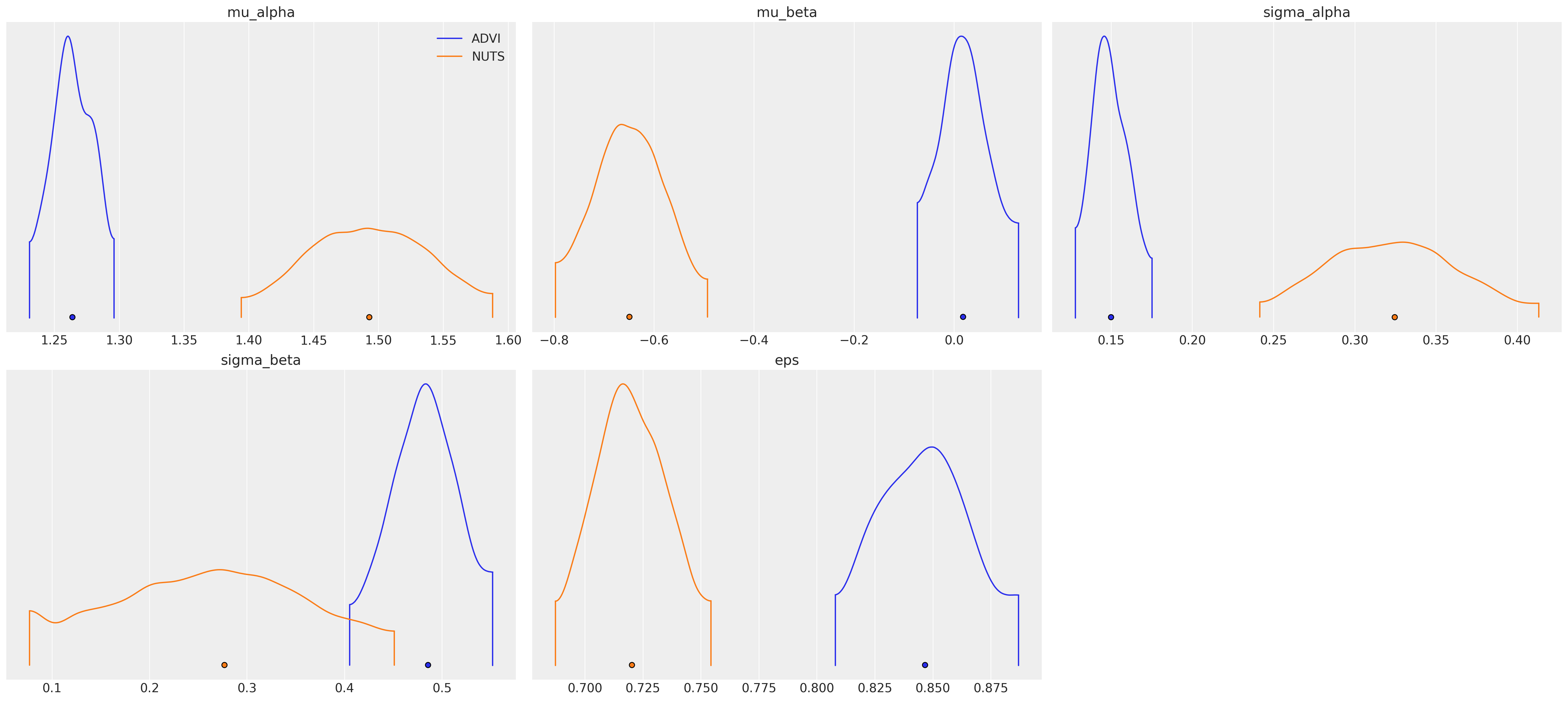
水印#
%load_ext watermark
%watermark -n -u -v -iv -w -p xarray
Last updated: Thu Sep 23 2021
Python implementation: CPython
Python version : 3.8.10
IPython version : 7.25.0
xarray: 0.17.0
numpy : 1.21.0
seaborn : 0.11.1
pandas : 1.2.1
matplotlib: 3.3.4
theano : 1.1.2
pymc3 : 3.11.2
scipy : 1.7.1
arviz : 0.11.2
Watermark: 2.2.0