进化一个种群#
在pygmo中,使用优化算法解决优化问题被描述为进化一个种群。在科学文献中,过去几十年里关于进化是否是一种优化形式的讨论非常有趣。在pygmo中,我们持相反的观点,认为所有类型的优化都是进化的一种形式。无论您使用的是SQP、内点优化器还是进化策略求解器,在pygmo中,您总是需要调用一个名为evolve()的方法来改进您的初始解,即您的种群。
进化种群的最简单方法是直接使用algorithm方法evolve()
>>> import pygmo as pg
>>> # The problem
>>> prob = pg.problem(pg.rosenbrock(dim = 10))
>>> # The initial population
>>> pop = pg.population(prob, size = 20)
>>> # The algorithm (a self-adaptive form of Differential Evolution (sade - jDE variant)
>>> algo = pg.algorithm(pg.sade(gen = 1000))
>>> # The actual optimization process
>>> pop = algo.evolve(pop)
>>> # Getting the best individual in the population
>>> best_fitness = pop.get_f()[pop.best_idx()]
>>> print(best_fitness)
[1.31392565e-07]
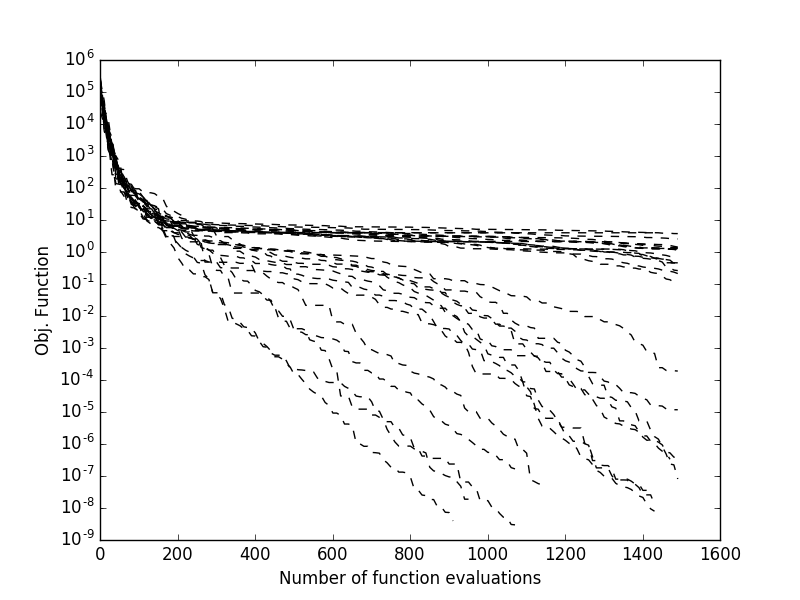
显然,由于sade是一种随机优化算法,如果我们从相同的种群开始重复进化,我们会得到不同的结果。如果我们想要监控来自不同初始种群的多次运行,并观察随着适应度评估次数的增加,最终最佳适应度是如何实现的。大多数pygmo UDA允许这样做,因为它们维护了一个内部日志,可以在UDA被提取后访问(参见extract())。这允许,例如,获得如右侧所示的图表,其中监控了多次试验:
>>> # We set the verbosity to 100 (i.e. each 100 gen there will be a log line)
>>> algo.set_verbosity(100)
>>> # We perform an evolution
>>> pop = pg.population(prob, size = 20)
>>> pop = algo.evolve(pop)
Gen: Fevals: Best: F: CR: dx: df:
1 20 135745 0.264146 0.541101 61.4904 2.55547e+06
101 2020 36.7951 0.252116 0.380181 7.01344 179.332
201 4020 4.06502 0.254534 0.740681 6.00596 59.3928
301 6020 2.96096 0.147331 0.790883 0.402499 7.95054
401 8020 2.43639 0.452624 0.922214 5.28217 6.80877
501 10020 1.65288 0.981585 0.975004 1.34442 0.897989
601 12020 1.16901 0.97663 0.783304 1.70733 1.36504
701 14020 0.750577 0.398881 0.922214 2.11136 1.78254
801 16020 0.438464 0.36723 0.960114 1.65512 1.18982
901 18020 0.18011 0.25882 0.932028 2.34781 1.4481
1001 20020 0.0792334 0.412775 0.887402 1.07089 0.211411
1101 22020 0.00875321 0.852287 0.472265 1.17808 0.170671
1201 24020 0.000820125 0.529034 0.633499 0.207262 0.0122566
1301 26020 2.13261e-05 0.176605 0.896106 0.060826 0.000706859
1401 28020 6.06571e-07 0.105885 0.81249 0.00852982 4.93967e-05
Exit condition -- generations = 1500
>>> uda = algo.extract(pg.sade)
>>> log = uda.get_log()
>>> import matplotlib.pyplot as plt
>>> plt.semilogy([entry[0] for entry in log],[entry[2]for entry in log], 'k--')
>>> plt.show()